Team:TU Delft/2 August 2010 content
From 2010.igem.org
(→Alkane degradation) |
|||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | =Alkane Sensing, Solvent Tolerance and Salt Tolerance= | + | =Lab work= |
| + | ==Alkane Sensing, Solvent Tolerance and Salt Tolerance== | ||
''by Pieter'' | ''by Pieter'' | ||
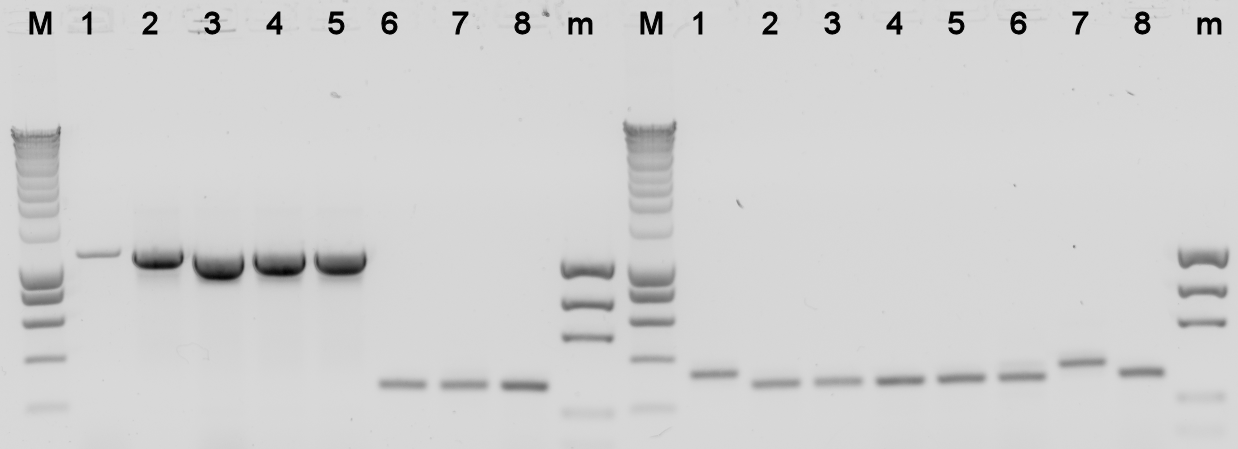
The plates containing yesterday's ligations contained colonies, to check whether they really contain the desired BioBrick a [[Team:TU_Delft/protocols/colony PCR|colony PCR]] was done, and the used colonies were grown in liquid LB medium over night. The results from the PCR were analysed on a 1% agarose gel. | The plates containing yesterday's ligations contained colonies, to check whether they really contain the desired BioBrick a [[Team:TU_Delft/protocols/colony PCR|colony PCR]] was done, and the used colonies were grown in liquid LB medium over night. The results from the PCR were analysed on a 1% agarose gel. | ||
| - | =Alkane degradation= | + | [[Image:TU_Delft_Pi2_Colony_PCR_part_1.png|400px]] |
| + | |||
| + | * BBa_K398305 = Alks -> E0240 | ||
| + | * BBa_K398101 = bbc1 -> J61101 | ||
| + | * BBa_K398402 = PhPFDalpha -> J61101 | ||
| + | * BBa_K398403 = PhPFDbeta -> J61101 | ||
| + | |||
| + | Lane Description: | ||
| + | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected Length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |M | ||
| + | |SmartLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |1 | ||
| + | |BBa_K398305 | ||
| + | |3772 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |BBa_K398305 | ||
| + | |3772 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |BBa_K398305 | ||
| + | |3772 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |BBa_K398305 | ||
| + | |3772 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |BBa_K398305 | ||
| + | |3772 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |BBa_K398101 | ||
| + | |1007 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |BBa_K398101 | ||
| + | |1007 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |8 | ||
| + | |BBa_K398101 | ||
| + | |1007 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |m | ||
| + | |EZ Ladder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |M | ||
| + | |SmartLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |1 | ||
| + | |BBa_K398101 | ||
| + | |1007 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |BBa_K398101 | ||
| + | |1007 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |BBa_K398402 | ||
| + | |836 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |BBa_K398402 | ||
| + | |836 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |BBa_K398402 | ||
| + | |836 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |BBa_K398402 | ||
| + | |836 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |BBa_K398402 | ||
| + | |836 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |8 | ||
| + | |BBa_K398403 | ||
| + | |734 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |m | ||
| + | |EZ Ladder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | [[Image:TU_Delft_Pi2_Colony_PCR_part_2.png|400px]] | ||
| + | |||
| + | Lane Description: | ||
| + | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected Length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |M | ||
| + | |SmartLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |1 - 7 | ||
| + | |Eva's samples | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |m | ||
| + | |EZ tLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |1 | ||
| + | |BBa_K398403 | ||
| + | |734 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |BBa_K398403 | ||
| + | |734 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |BBa_K398403 | ||
| + | |734 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |BBa_K398403 | ||
| + | |734 | ||
| + | |G00101 + G00100 | ||
| + | |<font color=red>X</font> | ||
| + | | | ||
| + | |- | ||
| + | |M | ||
| + | |SmartLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | ==Emulsifier== | ||
| + | When going over the plates in the fridge we noticed a few white colonies on plates from 13-07-'10. Previously we thought there were only red colonies. So we decided to do a [[Team:TU_Delft/protocols/colony PCR|colony PCR]] on all 12 white colonies. | ||
| + | |||
| + | [[Image:TU_Delft_Pi3_Colony_PCR.jpg|400px]] | ||
| + | |||
| + | *BBa_K398202 = R0011 + B0032 | ||
| + | |||
| + | Lane Description: | ||
| + | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected Length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |M | ||
| + | |SmartLadder | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | | | ||
| + | |- | ||
| + | |1 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |8 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |9 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | |Isolated plasmid | ||
| + | |- | ||
| + | |10 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |11 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |12 | ||
| + | |BBa_K398202 | ||
| + | |392 | ||
| + | |G00101 + G00100 | ||
| + | | | ||
| + | |Isolated plasmid | ||
| + | |} | ||
| + | |||
| + | None of the bands is truly positive. Still number 4,5 and 9 could be positive. Or due to the GC content the bands could be shifted a little bit upwards, so that maybe even sample 12 could be positive. | ||
| + | |||
| + | Just to be sure, we isolated the plasmids and send them to BaseClear for sequencing. The results will be in by Thursday. | ||
| + | |||
| + | ==Alkane degradation== | ||
For the next step in BioBrick formation a [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digestion]] was done: | For the next step in BioBrick formation a [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digestion]] was done: | ||
| Line 92: | Line 430: | ||
Results of the digestion on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]] | Results of the digestion on [[Team:TU_Delft/protocols/agarose_gel|1% agarose gel]] | ||
| - | [[Image:TUDelft_20100802_digestion.png|400px|1% Agarose of digestion check ]] | + | [[Image:TUDelft_20100802_digestion.png|400px|thumb|left|1% Agarose of digestion check ]] |
| - | gel runned at 100V for 1 hour | + | The gel was runned at 100V for 1 hour, loaded 5 μL of marker and 5 μL sample + 1 μL loadingbuffer |
| - | loaded 5 μL of marker and | + | |
Lane description: | Lane description: | ||
| Line 102: | Line 439: | ||
|'''Expected Length (bp)''' | |'''Expected Length (bp)''' | ||
|'''Status''' | |'''Status''' | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|- | |- | ||
|1 | |1 | ||
| - | | | + | |Smartladder |
| | | | ||
| - | |||
| | | | ||
|- | |- | ||
|2 | |2 | ||
| - | | | + | |‘E – J61100-AlkB2 – S’ (007T) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=red>✗</font> | ||
|- | |- | ||
|3 | |3 | ||
| - | |Undigested | + | |Undigested 007T |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=red>✗</font> | ||
|- | |- | ||
|4 | |4 | ||
| - | | | + | |‘E – J61100-rubA3 – X’ (008A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|5 | |5 | ||
| - | |Undigested | + | |Undigested 008A |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|6 | |6 | ||
| - | | | + | |‘E – J61100-rubR – S’ (010A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|7 | |7 | ||
| - | |Undigested | + | |Undigested 010A |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|8 | |8 | ||
| - | | | + | |‘E – B0015 – pSB1AK3 – X’ |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|9 | |9 | ||
| - | | | + | |pSB1AK3-B0015 |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|10 | |10 | ||
| - | | | + | |‘E – J61100-ladA – S’ (017A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|11 | |11 | ||
| - | |Undigested | + | |Undigested 017A |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|12 | |12 | ||
| - | | | + | |‘X – J61101-ADH – P’ (018A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|13 | |13 | ||
| - | |Undigested | + | |Undigested 018A |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|14 | |14 | ||
| - | | | + | |‘E – J61107-ALDH – S’ (019A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|15 | |15 | ||
| - | |Undigested | + | |Undigested (019A) |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|16 | |16 | ||
| - | | | + | |‘E – pSB1K3 – P’ |
| - | + | ||
| - | + | ||
| | | | ||
| + | |<font color=limegreen>✓</font> | ||
|- | |- | ||
|17 | |17 | ||
| - | | | + | |Smartladder |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| | | | ||
| - | |||
| | | | ||
|} | |} | ||
Latest revision as of 16:08, 15 August 2010
Contents |
Lab work
Alkane Sensing, Solvent Tolerance and Salt Tolerance
by Pieter
The plates containing yesterday's ligations contained colonies, to check whether they really contain the desired BioBrick a colony PCR was done, and the used colonies were grown in liquid LB medium over night. The results from the PCR were analysed on a 1% agarose gel.
- BBa_K398305 = Alks -> E0240
- BBa_K398101 = bbc1 -> J61101
- BBa_K398402 = PhPFDalpha -> J61101
- BBa_K398403 = PhPFDbeta -> J61101
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 2 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 3 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 4 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 5 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 6 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 7 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 8 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| m | EZ Ladder | n/a | n/a | n/a | |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 2 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 3 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 4 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 5 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 6 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 7 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 8 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| m | EZ Ladder | n/a | n/a | n/a |
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 - 7 | Eva's samples | n/a | n/a | n/a | |
| m | EZ tLadder | n/a | n/a | n/a | |
| 1 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 2 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 3 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 4 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| M | SmartLadder | n/a | n/a | n/a |
Emulsifier
When going over the plates in the fridge we noticed a few white colonies on plates from 13-07-'10. Previously we thought there were only red colonies. So we decided to do a colony PCR on all 12 white colonies.
- BBa_K398202 = R0011 + B0032
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 2 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 3 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 4 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 5 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 6 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 7 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 8 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 9 | BBa_K398202 | 392 | G00101 + G00100 | Isolated plasmid | |
| 10 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 11 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 12 | BBa_K398202 | 392 | G00101 + G00100 | Isolated plasmid |
None of the bands is truly positive. Still number 4,5 and 9 could be positive. Or due to the GC content the bands could be shifted a little bit upwards, so that maybe even sample 12 could be positive.
Just to be sure, we isolated the plasmids and send them to BaseClear for sequencing. The results will be in by Thursday.
Alkane degradation
For the next step in BioBrick formation a digestion was done:
| # | Sample | Enzyme 1 | Enzyme 2 | Enzyme 3 | Buffer | BSA | Needed fragment |
| 1 | 1 μg 007T | EcoRI | SpeI | BamH1 | 2 (Biolabs) | ✓ | ‘E – J61100-AlkB2 – S’ |
| 2 | 1 μg 008A | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘E – J61100-rubA3 – X’ | |
| 3 | 1 μg 010A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61100-rubR – S’ |
| 4 | 2 μg B0015 | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘E – B0015 – pSB1AK3 – X’ | |
| 5 | 1 μg 017A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61100-ladA – S’ |
| 6 | 1 μg 018A | XbaI | PstI | AseI | 2 (Biolabs) | ✓ | ‘X – J61101-ADH – P’ |
| 7 | 1 μg 019A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61107-ALDH – S’ |
| 8 | 1 μg pSB1K3 | EcoRI | PstI | 2 (Biolabs) | ✓ | ‘E – pSB1K3 – P’ |
Results of the digestion on 1% agarose gel
The gel was runned at 100V for 1 hour, loaded 5 μL of marker and 5 μL sample + 1 μL loadingbuffer
Lane description:
| # | Description | Expected Length (bp) | Status |
| 1 | Smartladder | ||
| 2 | ‘E – J61100-AlkB2 – S’ (007T) | ✗ | |
| 3 | Undigested 007T | ✗ | |
| 4 | ‘E – J61100-rubA3 – X’ (008A) | ✓ | |
| 5 | Undigested 008A | ✓ | |
| 6 | ‘E – J61100-rubR – S’ (010A) | ✓ | |
| 7 | Undigested 010A | ✓ | |
| 8 | ‘E – B0015 – pSB1AK3 – X’ | ✓ | |
| 9 | pSB1AK3-B0015 | ✓ | |
| 10 | ‘E – J61100-ladA – S’ (017A) | ✓ | |
| 11 | Undigested 017A | ✓ | |
| 12 | ‘X – J61101-ADH – P’ (018A) | ✓ | |
| 13 | Undigested 018A | ✓ | |
| 14 | ‘E – J61107-ALDH – S’ (019A) | ✓ | |
| 15 | Undigested (019A) | ✓ | |
| 16 | ‘E – pSB1K3 – P’ | ✓ | |
| 17 | Smartladder |
 "
"