Team:Stockholm/Planning
From 2010.igem.org
m (→IgG protease) |
m (→Practical schedule) |
||
| (11 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | == | + | ==Background== |
| Line 656: | Line 656: | ||
'''Johan''': Maybe there has been some tricks developed the last 5 years that we can take advantage of (anyone?), otherwise it seems that we have to use http://partsregistry.org/wiki/index.php?title=Part:BBa_J32015 and maybe get it to leak to the outside? (anyone disagree?). For Lactobacillus it seems to be ezmode, just add http://partsregistry.org/wiki/index.php?title=Part:BBa_K128004 | '''Johan''': Maybe there has been some tricks developed the last 5 years that we can take advantage of (anyone?), otherwise it seems that we have to use http://partsregistry.org/wiki/index.php?title=Part:BBa_J32015 and maybe get it to leak to the outside? (anyone disagree?). For Lactobacillus it seems to be ezmode, just add http://partsregistry.org/wiki/index.php?title=Part:BBa_K128004 | ||
| - | ===Choise of | + | ===Choise of chassis=== |
''"In iGEM, and synthetic biology in general, the microorganism that is being worked with is referred to as the cellular chassis (shăs'ē, chăs'ē). For most iGEM teams, the chassis will be E. coli, but you are not limited to working with E. coli in iGEM. Some of the past chassis have been Bacillus subtilis, lactobacillus, yeast, and even some mammalian cells. | ''"In iGEM, and synthetic biology in general, the microorganism that is being worked with is referred to as the cellular chassis (shăs'ē, chăs'ē). For most iGEM teams, the chassis will be E. coli, but you are not limited to working with E. coli in iGEM. Some of the past chassis have been Bacillus subtilis, lactobacillus, yeast, and even some mammalian cells. | ||
| Line 688: | Line 688: | ||
• Lactobacillus (Very friendly for the skin) | • Lactobacillus (Very friendly for the skin) | ||
| - | * | + | * https://2007.igem.org/Edinburgh/Yoghurt/Design#Gene_Expression_In_Lactobacillus |
* https://2008.igem.org/Team:MIT#Lactobacillus_Work | * https://2008.igem.org/Team:MIT#Lactobacillus_Work | ||
* http://openwetware.org/wiki/Category:Lactobacillus | * http://openwetware.org/wiki/Category:Lactobacillus | ||
| Line 702: | Line 702: | ||
'''Andreas''': Found some information on using Lactobacilli as our cloning organism for the project. | '''Andreas''': Found some information on using Lactobacilli as our cloning organism for the project. | ||
The Edinburgh 2007 iGEM team used a Lactobacillus strain (probably acidophilus?) for their yoghurt project. They created a vector which they successfully used in both Lactobacillus and E. coli. The vector is available as BBa_I742103 and the project link is attached below: | The Edinburgh 2007 iGEM team used a Lactobacillus strain (probably acidophilus?) for their yoghurt project. They created a vector which they successfully used in both Lactobacillus and E. coli. The vector is available as BBa_I742103 and the project link is attached below: | ||
| - | + | https://2007.igem.org/Edinburgh/Yoghurt/Design#Gene_Expression_In_Lactobacillus | |
I also found out that many Lactobacilli have pretty short generation times (down to 25 minutes), so that shouldn’t be a problem either. | I also found out that many Lactobacilli have pretty short generation times (down to 25 minutes), so that shouldn’t be a problem either. | ||
| Line 774: | Line 774: | ||
- Andreas Constantinou | - Andreas Constantinou | ||
- Emmelie Lidh | - Emmelie Lidh | ||
| - | |||
- Hassan Asl Foroughi | - Hassan Asl Foroughi | ||
| Line 824: | Line 823: | ||
'''Wet-lab''' | '''Wet-lab''' | ||
| - | * Nina: CPP, Protein A, | + | * Nina: CPP, Protein A, Tyrosinase, IgG protease |
| - | * Johan: CPP, | + | * Johan: CPP, bFGF |
| - | * Andreas: CPP, | + | * Andreas: CPP, SOD+yCSS |
| - | * Emmelie: CPP, MITF | + | * Emmelie: CPP, MITF |
'''Jamboree''' | '''Jamboree''' | ||
| Line 947: | Line 946: | ||
* GFP - Part:BBa_E0040 http://partsregistry.org/Part:BBa_E0040 | * GFP - Part:BBa_E0040 http://partsregistry.org/Part:BBa_E0040 | ||
| - | + | ==DNA sequences== | |
| - | === | + | ===What to look out for=== |
| - | ==== | + | ====Restriction sites==== |
* BBa_M31982 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31982, NotI restriction enzyme site, gcggccgc | * BBa_M31982 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31982, NotI restriction enzyme site, gcggccgc | ||
| Line 986: | Line 985: | ||
For details on why, check the article "Engineering BioBrick vectors from BioBrick parts", table 1. | For details on why, check the article "Engineering BioBrick vectors from BioBrick parts", table 1. | ||
| + | === CPP === | ||
| - | + | ==== Transportan 10 (Ülo Langel's) ==== | |
| - | + | ||
| - | + | ||
'''Amino acid sequence:'''<br> | '''Amino acid sequence:'''<br> | ||
| Line 1,001: | Line 999: | ||
- (http://genomes.urv.es/OPTIMIZER) | - (http://genomes.urv.es/OPTIMIZER) | ||
| - | + | '''Restriction sites:''' none | |
| - | + | ==== Low-molecular-weight protamine (LMWP) peptide ==== | |
'''Amino acid sequence:'''<br> | '''Amino acid sequence:'''<br> | ||
<tt>VSRRRRRRGGRRRR</tt> | <tt>VSRRRRRRGGRRRR</tt> | ||
| Line 1,014: | Line 1,012: | ||
- (http://genomes.urv.es/OPTIMIZER) | - (http://genomes.urv.es/OPTIMIZER) | ||
| - | + | '''Restriction sites:''' none | |
| - | + | ==== TAT ("Dowdy's CPP") ==== | |
'''Amino acid sequence:'''<br> | '''Amino acid sequence:'''<br> | ||
<tt>YGRKKRRQRRR</tt> | <tt>YGRKKRRQRRR</tt> | ||
| Line 1,027: | Line 1,025: | ||
- (http://genomes.urv.es/OPTIMIZER) | - (http://genomes.urv.es/OPTIMIZER) | ||
| - | + | '''Restriction sites:''' none | |
| - | + | === Protein A (Z domain)=== | |
| - | <tt> | + | <tt>GTAGACAAC'''AAATTC'''AACAAAGAACAACAAAACGCGTTCTATGAGATCTTACATTTACCT<br> |
AACTTAAACGAAGAACAACGAAACGCCTTCATCCAAAGTTTAAAAGATGACCCAAGCCAA<br> | AACTTAAACGAAGAACAACGAAACGCCTTCATCCAAAGTTTAAAAGATGACCCAAGCCAA<br> | ||
AGCGCTAACCTTTTAGCAGAAGCTAAAAAGCTAAATGATGCTCAGGCGCCGAAA</tt> | AGCGCTAACCTTTTAGCAGAAGCTAAAAAGCTAAATGATGCTCAGGCGCCGAAA</tt> | ||
| Line 1,036: | Line 1,034: | ||
- '''A synthetic IgG-binding domain based on staphylococcal protein A''', p. 107 | - '''A synthetic IgG-binding domain based on staphylococcal protein A''', p. 107 | ||
| - | + | '''Restriction sites:''' ApoI (10) | |
| - | + | === IgG protease === | |
<tt>ATGGATAGTTTTTCTGCTAATCAAGAGATTAGATATTCGGAAGTAACACCTTATCACGTT<br> | <tt>ATGGATAGTTTTTCTGCTAATCAAGAGATTAGATATTCGGAAGTAACACCTTATCACGTT<br> | ||
ACTTCCGTTTGGACCAAAGGAGTTACTCCTCCAGCAAACTTCACTCAAGGTGAAGATGTT<br> | ACTTCCGTTTGGACCAAAGGAGTTACTCCTCCAGCAAACTTCACTCAAGGTGAAGATGTT<br> | ||
| Line 1,060: | Line 1,058: | ||
Contains 1 AvrII & 1 Nsil, not so important | Contains 1 AvrII & 1 Nsil, not so important | ||
| - | + | === Vitamin D=== | |
- | - | ||
| - | + | === Vitamin B9 === | |
| - | + | ==== Vitamin B9 cluster ==== | |
<tt>>pNZ7026 pCon + folate gene cluster van plantarum<br> | <tt>>pNZ7026 pCon + folate gene cluster van plantarum<br> | ||
| Line 1,197: | Line 1,195: | ||
TTGGTTTTCTAATTTTGGTTCAAAGAAAGCTTGAGCTCTCTAGAACTAGTGGTAC</tt> | TTGGTTTTCTAATTTTGGTTCAAAGAAAGCTTGAGCTCTCTAGAACTAGTGGTAC</tt> | ||
| - | + | ==== FolP ==== | |
| - | + | ||
<tt>ATTTATGGCATCATGAATATTACGCCGGACTCCTTTTTCGACGGTGGCCAGTACAAGACT<br> | <tt>ATTTATGGCATCATGAATATTACGCCGGACTCCTTTTTCGACGGTGGCCAGTACAAGACT<br> | ||
AAAGATGACGTACTGAATCACGTGGATGCCATGTTACAAGCAGGAGCTGACGTAATCGAA<br> | AAAGATGACGTACTGAATCACGTGGATGCCATGTTACAAGCAGGAGCTGACGTAATCGAA<br> | ||
| Line 1,216: | Line 1,213: | ||
'''Restriction sites:''' XbaI (486), ApoI (559), MfeI (733) | '''Restriction sites:''' XbaI (486), ApoI (559), MfeI (733) | ||
| - | + | ==== Xtp2 ==== | |
<tt>ATGACGACGACTTGGTTGATTGCATCCAACAACGCTGGTAAAAGCCGCGACTTGATCGCG<br> | <tt>ATGACGACGACTTGGTTGATTGCATCCAACAACGCTGGTAAAAGCCGCGACTTGATCGCG<br> | ||
TGTTTGGCTTACTATGGCTTGACTGCTCGTCAGTACTTGACAGTGGCGCCGCGGCTCGAA<br> | TGTTTGGCTTACTATGGCTTGACTGCTCGTCAGTACTTGACAGTGGCGCCGCGGCTCGAA<br> | ||
| Line 1,230: | Line 1,227: | ||
'''Restriction sites:''' none | '''Restriction sites:''' none | ||
| - | + | ==== FolC2 ==== | |
<tt>GTGGATACAGGGACGATTGAACAACGTTATCAGGACTTATTAGCCCAACTAAATCAAGCC<br> | <tt>GTGGATACAGGGACGATTGAACAACGTTATCAGGACTTATTAGCCCAACTAAATCAAGCC<br> | ||
ATGTTAGCCAATAATCATCAGCGTGTTCCGTTATTACGGCGCATCATGGCACATCTTGGC<br> | ATGTTAGCCAATAATCATCAGCGTGTTCCGTTATTACGGCGCATCATGGCACATCTTGGC<br> | ||
| Line 1,257: | Line 1,254: | ||
'''Restriction sites:''' ApoI (301), NheI (358), SpeI (984) | '''Restriction sites:''' ApoI (301), NheI (358), SpeI (984) | ||
| - | + | ==== FolE ==== | |
<tt>ATGATTGATGAGAAGAACCAAGCAAAGATTGAGCATGCAGTACGAGAAATTTTAAGTGCA<br> | <tt>ATGATTGATGAGAAGAACCAAGCAAAGATTGAGCATGCAGTACGAGAAATTTTAAGTGCA<br> | ||
GTTGGTGAAGATCCTGACCGACCGGGATTAGTAGAAACACCAGCACGGGTTGCACGAATG<br> | GTTGGTGAAGATCCTGACCGACCGGGATTAGTAGAAACACCAGCACGGGTTGCACGAATG<br> | ||
| Line 1,271: | Line 1,268: | ||
'''Restriction sites:''' none | '''Restriction sites:''' none | ||
| - | + | ==== FolK ==== | |
<tt>ATGGCAAGTAGGGAAGAACGGGTTTATTTGAGTGTTGGTTCCAATATTCATCCGCGCGTC<br> | <tt>ATGGCAAGTAGGGAAGAACGGGTTTATTTGAGTGTTGGTTCCAATATTCATCCGCGCGTC<br> | ||
CAAAATATTCAGCAAGCCCTTAGCCGATTACGAGCCGTCAATGGGGTAAACGTGATTGAC<br> | CAAAATATTCAGCAAGCCCTTAGCCGATTACGAGCCGTCAATGGGGTAAACGTGATTGAC<br> | ||
| Line 1,284: | Line 1,281: | ||
'''Restriction sites:''' none | '''Restriction sites:''' none | ||
| - | + | ==== FolB ==== | |
<tt>ATGGGCATGATTCGAATTAATAATTTACGCTTTCACACGTTTAACGGGGTACTTCCGGAA<br> | <tt>ATGGGCATGATTCGAATTAATAATTTACGCTTTCACACGTTTAACGGGGTACTTCCGGAA<br> | ||
GAACGGCGTAATGGTCAACAACTAGGGCTAGATATTGCCATTAAATATCCTATCGAAACC<br> | GAACGGCGTAATGGTCAACAACTAGGGCTAGATATTGCCATTAAATATCCTATCGAAACC<br> | ||
| Line 1,295: | Line 1,292: | ||
'''Restriction sites:''' NheI (222) | '''Restriction sites:''' NheI (222) | ||
| - | + | ===Tyrosinase=== | |
DNA Sequence - http://partsregistry.org/wiki/index.php?title=Part:BBa_K193600<br> | DNA Sequence - http://partsregistry.org/wiki/index.php?title=Part:BBa_K193600<br> | ||
| Line 1,334: | Line 1,331: | ||
'''Restriction sites: ''' NsiI (1035), NgoMIV (1317) | '''Restriction sites: ''' NsiI (1035), NgoMIV (1317) | ||
| - | + | ===Catalase=== | |
- | - | ||
| - | + | ===Superoxidase dismutase=== | |
(dimer without unspecific crosslinking) | (dimer without unspecific crosslinking) | ||
| - | + | ====C6A & C11A (crosslinking)==== | |
<tt>ATGGCGACGAAGGCCGTGGCCGTGCTGAAGGGCGACGGCCCAGTGCAGGGCATCATCAAT<br> | <tt>ATGGCGACGAAGGCCGTGGCCGTGCTGAAGGGCGACGGCCCAGTGCAGGGCATCATCAAT<br> | ||
TTCGAGCAGAAGGAAAGTAATGGACCAGTGAAGGTGTGGGGAAGCATTAAAGGACTGACT<br> | TTCGAGCAGAAGGAAAGTAATGGACCAGTGAAGGTGTGGGGAAGCATTAAAGGACTGACT<br> | ||
| Line 1,352: | Line 1,349: | ||
'''Restriction sites:''' none | '''Restriction sites:''' none | ||
| - | + | ====yCCS (yeast copper chaperone)==== | |
<tt>ATGACCACGAACGATACATACGAGGCTACTTATGCCATTCCCATGCACTGTGAAAATTGC<br> | <tt>ATGACCACGAACGATACATACGAGGCTACTTATGCCATTCCCATGCACTGTGAAAATTGC<br> | ||
GTGAATGATATAAAGGCATGCCTAAAAAATGTCCCAGGAATCAATTCATTGAATTTCGAT<br> | GTGAATGATATAAAGGCATGCCTAAAAAATGTCCCAGGAATCAATTCATTGAATTTCGAT<br> | ||
| Line 1,369: | Line 1,366: | ||
'''Restriction sites: ''' EcoRI (228) | '''Restriction sites: ''' EcoRI (228) | ||
| - | + | ===bFGF=== | |
<tt>ATGGCAGCCGGGAGCATCACCACGCTGCCCGCCTTGCCCGAGGATGGCGGCAGCGGCGCC<br> | <tt>ATGGCAGCCGGGAGCATCACCACGCTGCCCGCCTTGCCCGAGGATGGCGGCAGCGGCGCC<br> | ||
TTCCCGCCCGGCCACTTCAAGGACCCCAAGCGGCTGTACTGCAAAAACGGGGGCTTCTTC<br> | TTCCCGCCCGGCCACTTCAAGGACCCCAAGCGGCTGTACTGCAAAAACGGGGGCTTCTTC<br> | ||
| Line 1,381: | Line 1,378: | ||
'''Restriction sites:''' AgeI (344) | '''Restriction sites:''' AgeI (344) | ||
| - | === | + | ===MGF=== |
| - | + | ||
| - | + | ||
| - | + | ||
- | - | ||
| + | |||
| + | ===MITF=== | ||
| + | GenBank: Z29678.1 | ||
| + | |||
| + | <tt>>gi|468496:121-1380 H.sapiens mitF mRNA<br> | ||
| + | ATGCTGGAAATGCTAGAATATAATCACTATCAGGTGCAGACCCACCTCGAAAACCCCACC<br> | ||
| + | AAGTACCACATACAGCAAGCCCAACGGCAGCAGGTAAAGCAGTACCTTTCTACCACTTTA<br> | ||
| + | GCAAATAAACATGCCAACCAAGTCCTGAGCTTGCCATGTCCAAACCAGCCTGGCGATCAT<br> | ||
| + | GTCATGCC'''ACCGGT'''GCCGGGGAGCAGCGCACCCAACAGCCCCATGGCTATGCTTACGCTT<br> | ||
| + | AACTCCAACTGTGAAAAAGAGGGATTTTATAAGTTTGAAGAGCAAAACAGGGCAGAGAGC<br> | ||
| + | GAGTGCCCAGGCATGAACACACATTCACGAGCGTCCTGTATGCAGATGGATGATGTAATC<br> | ||
| + | GATGACATCATTAGCCTAGAATCAAGTTATAATGAGGAAATCTTGGGCTTGATGGATCCT<br> | ||
| + | GCTTTGCAAATGGCAAATACGTTGCCTGTCTCGGGAAACTTGATTGATCTTTATGGAAAC<br> | ||
| + | CAAGGTCTGCCCCCACCAGGCCTCACCATCAGCAACTCCTGTCCAGCCAACCTTCCCAAC<br> | ||
| + | ATAAAAAGGGAGCTCACAGCGTGTATTTTTCCCACAGAGTCTGAAGCAAGAGCACTGGCC<br> | ||
| + | AAAGAGAGGCAGAAAAAGGACAATCACAACCTGATTGAACGAAGAAGAAGATTTAACATA<br> | ||
| + | AATGACCGCATTAAAGAACTAGGTACTTTGATTCCCAAGTCAAATGATCCAGACATGCGC<br> | ||
| + | TGGAACAAGGGAACCATCTTAAAAGCATCCGTGGACTATATCCGAAAGTTGCAACGAGAA<br> | ||
| + | CAGCAACGCGCAAAAGAACTTGAAAACCGACAGAAGAAACTGGAGCACGCCAACCGGCAT<br> | ||
| + | TTGTTGCTCAGAATACAGGAACTTGAAATGCAGGCTCGAGCTCATGGACTTTCCCTTATT<br> | ||
| + | CCATCCACGGGTCTCTGCTCTCCAGATTTGGTGAATCGGATCATCAAGCAAGAACCCGTT<br> | ||
| + | CTTGAGAA'''CTGCAG'''CCAAGACCTCCTTCAGCATCATGCAGACCTAACCTGTACAACAACT<br> | ||
| + | CTCGATCTCACGGATGGCACCATCACCTTCAACAACAACCTCGGAACTGGGACTGAGGCC<br> | ||
| + | AACCAAGCCTATAGTGTCCCCACAAAAATGGGATCCAAACTGGAAGACATCCTGATGGAC<br> | ||
| + | GACACCCTTTCTCCCGTCGGTGTCACTGATCCACTCCTTTCCTCAGTGTCCCCCGGAGCT<br> | ||
| + | TCCAAAACAAGCAGCCGGAGGAGCAGTATGAGCATGGAAGAGACGGAGCACACTTGTTAG</tt> | ||
| + | |||
| + | '''Restriction sites:''' AgeI (189), PstI (969) | ||
===Rare codons=== | ===Rare codons=== | ||
| Line 1,429: | Line 1,451: | ||
Note! We will use iGEM team Freiburg 2007 fusion parts (BBF RFC 25) as our assembly standard 25 and thus their prefix and suffix, which differ from the standard prefix and suffix. | Note! We will use iGEM team Freiburg 2007 fusion parts (BBF RFC 25) as our assembly standard 25 and thus their prefix and suffix, which differ from the standard prefix and suffix. | ||
| - | + | https://2007.igem.org/Freiburg07/report_fusion_parts | |
http://openwetware.org/wiki/The_BioBricks_Foundation:Standards/Technical/Formats#The_Berkeley_.28BBb.29_Format scroll down to BBF RFC 25. | http://openwetware.org/wiki/The_BioBricks_Foundation:Standards/Technical/Formats#The_Berkeley_.28BBb.29_Format scroll down to BBF RFC 25. | ||
Latest revision as of 12:30, 12 August 2010
Background
Here follows the ongoing discussion amongst the group members about the project we are putting together. In addition we include theoretical support from scientific articles for what we want to accomplish.
Cell penetrating peptide (CPP)
(High importance, really needs to be adressed. What is the best approach to penetrate the skin? What is the best approach to enter target cells? Can one approach solve both problems? If not, would the two approaches interfere with each other?)
"Protein drugs, because of their large size and hydrophilic nature, are normally precluded from effective delivery such as cell entry or tissue diffusion. Among the transport barriers, the skin poses a formidable challenge to proteins because of the impermeable stratum corneum." - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: I found this interesting article about a molecule that can penetrate the skin. Thus we have to build fusion proteins of our protein of interest together with a molecule that can penetrate the skin . This is to have them passing through the stratum corneum, which as the article says is a very good barrier.
"To circumvent these problems that confront the current methods, we hypothesized the concept of “skin-permeable proteins”, which would possess skin-penetrating ability and thereby eliminate the need for a transport vehicle." - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: The skin-permeable proteins seem to be good for our study concerning a model to have our proteins penetrating the skin.
"[...] we present a novel strategy for chemically constructing artificial skin-permeable proteins, illustrated by the simple conjugation of a protein to a cell-penetrating peptide (CPP), which would display a penetration effect on the stratum corneum barrier and transport the attached proteins into the skin." -Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: They have chemically constructed their CPP, but we will use a recombinant approach, meaning we will insert our genes of interest into plasmids and have bacteria expressing our proteins.
"The CPPs are capable of transporting their cargos, often linked by a covalent bond, into almost all cell types.[2] Among such CPPs, the low-molecular-weight protamine (LMWP) peptide (VSRRRRRRGGRRRR), developed in our laboratory by enzymatic digestion of protamine (an FDA-approved drug)" - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: Their CPP called LMWP has many arginine (R), thus the peptide is very positively charged making it easy to go through lipid layers.
"Secondly, unlike other CPPs, the toxicity profile of LMWP has already been thoroughly established. LMWP was shown to be nonimmunogenic,[4] and its use in dogs did not elicit acute toxic responses.[5] Lastly, while other CPPs must be chemically synthesized, LMWP can be produced in mass quantities direct from native protamine with limited processing time and cost.[6]" - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: It is good for us that LMWP has already been shown to be nonimmunogenic and also does not give toxic responses.
"[...] skin keratinocytes are a physical barrier that provides the front line of defense against infection and also poses a challenge to protein delivery." - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p. 2724
Nina: We must look further into the keratinocytes. How fast is their turn over, this is good to know when inserting any molecules into them.
"LMWP was shown to exhibit an ability to translocate linked cargos of varying sizes into keratinocytes, thus demonstrating the potential for percutaneous protein delivery." - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p.2725
Nina: We should have in mind the sizes of the proteins and other molecules we want to insert into the skin - everything has a limit.
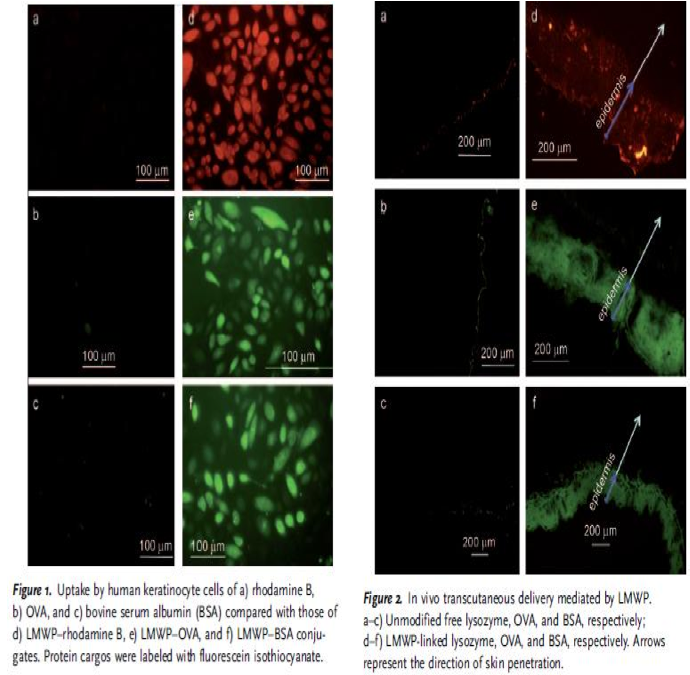
Nina: I added this picture from the article in order to show how they have demonstrated their CPP bound to the proteins and that they have entered both keratinocytes, but also the epidermis of the skin, which is very good for us that someone has already shown this.
"The plausibility of percutaneous delivery in vivo was examined by topical application of LMWP-linked lysozyme, OVA, or bovine serum albumin (BSA), to represent a broad range of protein sizes. All the LMWP-linked proteins successfully penetrated the stratum corneum and accumulated primarily in the epidermis (Figure 2), whereas the control proteins without LMWP linkage remained on the surface of the skin." - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization p.2725
Nina: This experiment is good in the way that it shows that big protein shuch as BSA (66 kDa) and OVA (45 kDa)fused with CPP can pass via skin.
“Since small basic proteins like protamines are precipitated by SDS, as an assay method we used acidic gels in which basic proteins migrate not only according to their size but also according to their charge. Thus, although small basic proteins can be identified in acidic gels, size calibration of these proteins is not accurate. Identification of specific proteins can be made by using deletion or other mutants to eliminate or change the nature of these proteins.” - An E. coli Gene Coding for a Protamine-Like Protein
Nina: I found this article since I wanted to know more about bacteria expressing protamines and their practical studies. So maybe think about using acidic gels when characterizing protamines, such as in our CPP.
“[...] that can code for a small basic protein directs the synthesis of such a protein in vitro. (We now define the P-protein coding region as the tpr locus for “tRNA followed by protamine-like protein.“) Mutants that lack part of the DNA region coding for the protein fail to direct its synthesis” - An E. coli Gene Coding for a Protamine-Like Protein
“In the first protocol, penetratin is synthesized with an additional N-terminalactivated cysteine, protected by a nitropyridinium group to prevent peptide homodimerization. The cargo is probably released freely in the cytoplasm as a result of disulfide bond breakage in the reductive cytoplasmic milieu. In the second method, the cargo and the vector are chemically synthesized in continuity. In this case, no coupling reaction is necessary.” - Handbook of Cell-Penetrating Peptides p 12 Ülo
Nina: This citation and the following three is interesting since they are focused on CPP. We should read the book in order to get a greater understanding about the CPPs.
It is interesting to know that the cargo can be release in a reductive cytoplasmic environment, thus the disulfides bonds will be reduced.
“Despite certain hydrophobicity provided by proline, the most important advantage of proline-rich peptides in biological systems is their high degree of water solubility, an essential property for their use in life sciences. [...] In general, it is accepted that CPPs offer several advantages over other known cellular delivery systems, including low toxicity, high efficiency towards distinct cell lines, and even inherent therapeutic potential.” - Handbook of Cell-Penetrating Peptides p 62 Ülo
Nina: Some CPPs seem to be somewhat well studied, now we maybe should find a couple of them that could be good in our project.
“[...] the metabolic degradation of the CPP and the CPP cargo is necessary for the delivery of the cargo after internalization and the final elimination of the CPP to avoid chronic toxicity. An equilibrium between these two aspects is required in order to avoid the premature release of the cargo and its cleavage once internalized.” - Handbook of Cell-Penetrating Peptides p 63 Ülo
Nina: We should learn more about the CPP degradation in order to understand and handle toxic levels.
“[...] permeability can be modulated by taking advantage of the amino side chain, which, in this case, is the a-amino function of the proline. Thus, through this side chain, several parameters of the CPP can be altered, such as hydrophilicity/hydrophobicity characteristics.” - Handbook of Cell-Penetrating Peptides p 63-64 Ülo
Nina: Maybe this information about this amino side chain is good for us to know during our practicals if we want to modulate this part.
Steven Dowdy (mentioned by Ülo Langel when discussing CCP using recombinant approach)
- TAT transduction: the molecular mechanism and therapeutic prospects (2007)
Johan: A lot of interesting information how CCPs seems to work (see all the yellowed text in Dropbox)
Nina: Dowy is very important for us since not many from what I have seen have used recombinant approach when synthesizing and fusing their CPPs with proteins except him.
- Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer (2005)
Johan: Similar to the first article but older
- Recent advances in the use of protein transduction domains for the delivery of peptides, proteins and nucleic acids in vivo (2005)
Johan: Soooo many examples of in-vivo experiments with CCPs
- Modulation of Cellular Function by TAT Mediated Transduction of Full Length Proteins (2003)
Johan: Similar to the first article but older
- Protein Transduction: Generation of Full-Length Transducible Proteins Using the TAT System (2003)
Johan: Talks about using CCPs with recombinant approach (what we will use. Most other articles expresses CCP and cargo seperately and then fuses them aided by some chemicals). Nina and Andreas, he talk about an Appendix, for example to read more about “further background into creating fusion cDNA”. Does anyone know how to find this appendix? I cant :(
- Cell Penetrating Peptides in Drug Delivery (2003)
Johan: Similar to the first article but older
Johan: (6 articles of him that I downloaded, of his ~90 articles in total). Seems that this field is going forward rapidly and that we should only focus on new articles (preferably newer than 2007 so they include data not present in the first article)
Protein A
Protein A would be secreted from the bacteria, penetrating the skin and bind IgG antibodies - that in vitiligo patients bind melanocytes - so that the igG antibodies can not induce an immune response.
"It binds proteins from many of mammalian species, most notably IgGs" - http://en.wikipedia.org/wiki/Protein_A (Find better source)
Johan: Good as IgGs are the main antibody in excess for vitiligo
“It binds with the Fc region of immunoglobulins through interaction with the heavy chain. The result of this type of interaction is that, in serum, the bacteria will bind IgG molecules in the wrong orientation (in relation to normal antibody function) on their surface which disrupts opsonization and phagocytosis.” - http://en.wikipedia.org/wiki/Protein_A
Johan: The disruption is what we want (are there more disruptions to think of, other than opsonization and phagocytosis?)
Nina: Those are the most important ones, which is very good that happens in our case. Later would be if we could combine this with an IgG protease that would destroy the auto-antibody IgG.
“It binds with high affinity to human IgG1 and IgG2 as well as mouse IgG2a and IgG2b. Protein A binds with moderate affinity to human IgM, IgA and IgE as well as to mouse IgG3 and IgG1.[1] It does not react with human IgG3 or IgD, nor will it react to mouse IgM, IgA or IgE.” - http://en.wikipedia.org/wiki/Protein_A
Johan: See if the antibody in excess in vitiligo hopefully is IgG1/IgG2
Nina: The other antibodies are not in any excess so we do not need to put much research on them, instead learn more about IgG.
“... possesses several properties which makes it suitable as a partner in fusion proteins: (a) SpA enables purification of the fusion protein by affinity chromatography due to its specific binding to the Fc part of immunoglobulins, (b) it is competent for secretion in E. coli, (c) it is monomeric and relatively small, (d) it has been reported to be extremely soluble in water solutions (Samuels- son et al., 1991), (e) it is proteolytically very stable intracellularly (Nilsson et al., 1985a) and in the periplasm of E. coli (Nilsson and Abrahms6n, 1990). Fusions to the IgG-binding domains of SpA have successfully been applied for high-level production of peptide hormones (Abrahms6n et al., 1986; Moks et al., 1987a, b; Nilsson et al., 1991, 1985b) and as a tool for the production of specific antibodies against gene products (L6wenadler et al., 1987, 1986).” - Fusions to the 5' end of a gene encoding a two-domain analogue of staphylococcal protein A, pp 270-271
Nina: This information is very good and gives a lot of hope on this protein.
Johan: There are shortened versions of protein A. These might have some advantages. E.g. they dont have the domain that normally binds to the cell membrane, instead they are secreted extracellularly.
Nina: Also it is very good that it is has become very small. :D
http://johan.nordholm.se/Skola/igem/Z.pdf Interesting information about the Z domain of Protein A, text from the doctoral thesis "Interaction-Engineered Three-Helix Bundle Domains for Protein Recovery and Detection" by Tove Alm from 2010 at KTH
http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=2SPZ
Andreas: How will we prevent protein A from entering the blood stream? If it is able to penetrate the skin cells to the spot where antibodies are present, it is also likely to ”exit” the skin into the blood stream. If this happens, protein A will not only cause a severe immune shock, it will also interfere with any type of antibody in the blood, not just the ones targeting vitiligo. This is especially true if Protein A is proteolytically very stable, as stated in (e) above.
Are there any examples mammalian in vivo Protein A studies? It would be a great leap forward if there are any such examples, showing how Protein A interacts with the mammalian immune system. - I found a website at the US National Cancer Insitute, where they mention two clinical trials aiming at using Staphylococcus aureus Protein A as a cancer treatment. The clinical trials made it to phase 2 (out of the usual 4, if I remember correctly), but were then closed. I haven’t been able to find any statements, links or articles on why they were closed, but here is a link to the NCI website: http://ncit.nci.nih.gov/ncitbrowser/ConceptReport.jsp?dictionary=NCI%20Thesaurus&code=C17166 - Now I found an article from the mid-80s on Protein A. Not sure if it is connected to the clinical trials at NCI, but here’s a PubMed link: http://www.ncbi.nlm.nih.gov/pubmed/6379115 - There are also several articles mentioning ex vivo methods with Protein A for cancer treatments.
Nina: Size of protein A with Z domain: 7 kDA
IgG protease
IgG antibodies are enriched in vitiligo patients. IgG proteases could partially solve this problem. Also, IgG protease could be fused with Protein A to get more activity from the protease.
"Actinobacillus actinomycetemcomitans produces a protease to human immunoglobulin G that is an important evasion mechanism […] The results showed that A. actinomycetemcomitans produced a protease to human immunoglobulin G in the culture supernatant […] The molecular mass of the protease active fraction was from 43 to 150 kDa." - IMMUNOGLOBULIN G PROTEOLYTIC ACTIVITY OF ACTINOBACILLUS ACTINOMYCETEMCOMITANS
Johan: Their IgG protease eluted in fraction 13-17. +150 kDa eluted in fraction 9, and 43 kDa eluted in fraction 19. So at least it seems that the IgG protease is closer to 43 kDa than to +150 kDa.. (I wouldnt want to be a CPP carrying a cargo +150 kDa...)
"IdeS, a proteinase from Streptococcus pyogenes, cleaves immunoglobulin (Ig)G antibodies with a unique degree of specificity. Pathogenic IgG antibodies constitute an important clinical problem contributing to the pathogenesis of a number of autoimmune conditions and acute transplant rejection. To be able to effectively remove such antibodies is therefore an important clinical challenge." - IdeS: A Bacterial Proteolytic Enzyme with Therapeutic Potential
Tyrosinase (produces melanin)
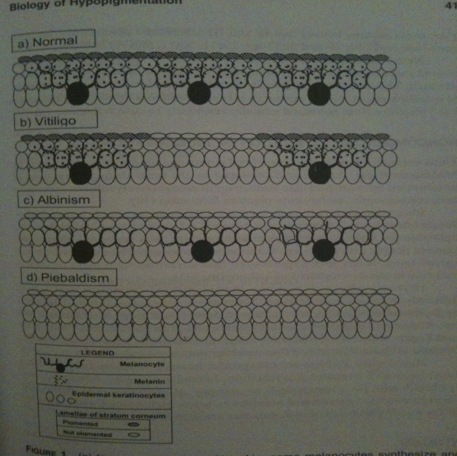
Nina: In the “normal“ skin melanocytes are present and secrete out melanin molecules which enter the keratocytes, resulting in color in the skin. In “vitiligo“ skin melanocytes are missing at some areas, thus no melanin production takes place at these patches that keratocytes can take up and result in skin color, instead one gets white patches. Note! Ignore albinism and piebaldism.
Nina: I scanned this picture from the book: Vitiligo Problems and solutions by Torello Lotti and Jana Hercogová. It is good in the way that is presents the relationship between keratinocytes and melanocytes. They are in close contact and have "communicate" with each other via molecules. In addition, it is shown here that melanocytes are like neronal cell with dendrite, from which they secrete molecules, such as melanin, to the keratinocytes. When melanocytes will be removed as in the vitiligo disease, they won't be able to secrete any necessary molecules to the keratinocytes such as melnin, which gives skin color. From here I thought that we could via our bacteia on the skin introduce the lacking melanin to the keratinocytes, hence in a short time period giving skin color. :)
Nina: Our aim is to have our bacteria on the skin to produce the missing melanin in vitiligo skin as a fusing protein with a cell penetrating peptide, thus passing into the skin layer (epidermis) and entering the “empty of melanin“ keratocytes. This will result in giving a vitiligo skin back its color somewhat directly (matter of days maybe, depending on how much melanin our bacteria produces and enters the target cells ->keratocytes). This approach of allowing for repigmentation to occur will help many people in a short time (compared to current techniques, taking 6 -12 months). Additionally, the vitiligo skin will be more tolerable against UV-light thanks to the bacteria produced melanin entering keratocytes.
Nina: We have found an article about transgenic e.coli producing one type of melanin. Hopefully in the future one might by research find a color spectrum of different pigment molecules (melanin) that will match to different people’s skin color.
“Melanins are polyphenolic heteropolymers with a wide range of colors and applications. Due to their chemical composition, melanins have physicochemical properties that allow them to act as ultraviolet absorbers, cation exchangers, drug carriers, amorphous semiconductors, X-ray and γ-ray absorbers, and in some instances, substrates with antioxidant and antiviral activity (Riley 1997; Lagunas-Muñoz et al. 2006). Bacteria are known to produce 3,4-dihydroxyphenylalanine-melanins, the pheomelanins or eumelanins, and melanin-like pigments that are derived from non-nitrogenous phenols (the allomelanins or pyomelanins; Gibson and George 1998).” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125
Nina: We should focus on the eumelanin. Remember these are not proteins, instead heteropolymers.
“Production of melanins by recombinant E. coli is useful in industrial processes.” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125
Nina: This article is good for us since we need to know that someone before have expressed melanin in e.coli and have succeded.
“The clone fss6 producing red-brown pigment was isolated and further characterized in the study. Based on the physico-chemical characteristics of the pigment, the red-brown pigment was identified as melanin.” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
"[...] the production of melanin was correlated with homogentistic acid (HGA). The p-hydroxyphenylpyruvate produced by the Escherichia coli host was converted to HGA through the oxidation reaction of introduced HPPD." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 124
Nina: Some pathways of melanin production, good to know more about.
"HPLC analysis for HGA from the cell-free filtrate of cultures showed that HGA was produced by recombinant E.coli strains before melanin was produced....HGA is the product of ORF2 and should be the precursor of melanin." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
Nina: Some pathways of melanin production, good to know more about.
"The cloned gene (ORF2) encoding putative 4-hydroxyphenylpyruvate dioxygenase (HPPD) was responsible for the production of melanin in E. coli." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
"The known pathway for the catabolism of tyrosine to HGA in mammals and bacteria is that tyrosine is converted to p-hydroxyphenylpyruvate (p-HPP) through a transamination reaction. Then, p-HPP is converted to HGA through the oxidation of HPPD (Lindstedt and Odelhög 1987; Lindstedt et al. 1977). In the study, the transaminase for the initial step should exist in clone fss6. Three aminotransferase capable of converting tyrosine into p-HPP have been identified in E. coli (Pittard 1987). We propose that tyrosine is first catabolized by aminotransferases of E. coli host, then, the produced p-HPP is converted to HGA by the introduced HPPD gene products. Lastly, the accumulation and polymerization of HGA results in the formation of melanin." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 130
Andreas: I find this part quite interesting and will look more into it. Since melanins are very small molecules, I’m not sure about using CPP as a shuttle into the cells, but since you say that melanin is produced by melanocytes, secreted and then enters the keratocytes, our bacterial melanin might be able to enter the keratocytes the same way.
Johan: Melanin is stored in organelles called melanosomes, this is a problem I’m looking into.
Nina: Mats found this part with the melanin very interesting and he told me to continue the melanin work by studying “eumelanin”.
- The biology of melanocytes
Johan: Has tons of interesting information, too much to write here (look how much is yellowed in the article in Dropbox)
”Our group has cloned the tyrosinase coding gene (melA) from the nitrogen-fixing bacterium Rhizobium etli CFN42 into the inducible vector pTrc99A and trans- formed it into E. coli W3110. This recombinant strain synthesizes EuMel in the presence of tyrosine and copper (Cabrera-Valladares et al. 2006). The present study was carried out to optimize tyrosine conversion into EuMel using melanogenic E. coli whole cells as biocatalyst in bench scale fermentors.” - Optimum melanin production using recombinant Escherichia coli, p .2
Nina: This article is very good and we will also need the melA gene coding for tyrosinase to have in a plasmid inserted into our bacteria.
Johan: This is good to hear. They just added extra tyrosine as powder and got a lot of melanin (under good conditions - 30° C pH 7). I can have missed, but I also didnt read that cells died, which is good, as the previous article mentions that “melanogenesis is hazard for melanin-producing cells” and that that is a reason why it is made in melanosomes.
“Furthermore, the disulfide linkage could be cleaved by the elevated level of glutathione and reductase activity in the cytosol,[12] which allowed release of OVA from LMWP and thus retention of a full intrinsic immunogenicity” - Synthetic Skin-Permeable Proteins Enabling Needleless Immunization pp.2725-2726
Johan: Dont fully understand the paragraph in the article, but it seems that the cargo could release from the CPP. This sounds very good as melanin exists in polymers and melanin+CPP would probably not be able to polymerize
Nina: Size of melanin: ~400 g/mol
MITF (Microphthalmia-associated transcription factor)
"MITF is a transcription factor that normal melanocytes secrete which acts beneficially towards the melanocyts, as mentioned below in the citations from an article. Vitiligo skin has unfortunately a disorder in producing MIFTs from their melanocytes. Our aim is to have our bacteria on the skin to produce MIFT as a fusion protein with a cell penetrating peptide, thus going thorugh the skin and protecting the melanocytes even if they naturally cannot produce MITF themselves. Although the biochemical pathway from tyrosine to the melanin polymer has been known for decades (1,2), the findings of the last fifteen years have led to unveiling the transcriptional regulation of genes which are necessary for the synthesis and deposition of melanin. Remarkably, only one transcription activator emerged as a universal regulator of expression of several proteins accomplishing the assembly of the melanosome and its decoration with melanin." (This protein --> MITF) - ‘‘Transcription physiology’’ of pigment formation in melanocytes: central role of MITF p. 2
Nina: MITF is mentioned a lot in articles such as this in the context of vitiligo. Therfore, I think this transcription factor seems to be good to introduce into the skin and further into melanocytes.
"Recently, beyond the already large group of genes activated by MITF, other potential targets were identified by using the microarrays in MITF-overexpressing human melanoma cells (20). Because severe MITF mutations preclude the development of embryonic melanocytes (21) and MITF is required for the survival of adult and even malignant melanocytes (22), the MITF protein is believed to be an essential regulator of the life and differentiation of melanocytes.
Melanomas generally express high levels of MITF, although these levels differ greatly among melanoma cell lines and cells in the tumor tissue." - ‘‘Transcription physiology’’ of pigment formation in melanocytes: central role of MITF p. 2
Nina: MITF seems to activate genes that code for enzymes which can take care of free radicals (ROS). The ROS have been shown to be accumulated in vitiligo skin since maybe a lack of these important enzymes, hence melanocytes die because of the many ROS.
"MITF controls not only expression of pigmentation-related genes but also genes involved in diverse biological processes in melanoma cells (Fig. 1a) such as proliferation, invasiveness, resistance to apoptosis and stress mediated by reactive oxygen species, and possibly metastasis." - ‘‘Transcription physiology’’ of pigment formation in melanocytes: central role of MITF p. 2
Nina: MITF looks to be very important to melanocytes.
"At least four different transcription factors participate in the transactivation of the MITF gene in melanocytes: the paired box-containing transcription factor PAX3, a sex determining region Y (SRY) family member SOX10, the Wnt ⁄ b-catenin pathway effector LEF-1 and the cAMP pathway effector cAMP response element binding (CREB) (Fig. 2), with the last two proteins being probably more important for the maintenance of MITF levels in melanoma cells" - ‘‘Transcription physiology’’ of pigment formation in melanocytes: central role of MITF p. 3
Nina: If it becomes to difficult in the practicals to introduce the transcription factor MITF, we may introduce instead the transcription factors involved in activating MITF in melanocytes, but hopefully we do not need to make this step.
"Isoform M is exclusively expressed in melanocytes and melanoma cells. Isoform A and isoform H are widely expressed in many cell types including melanocytes and retinal pigment epithelium (RPE). Isoform C is expressed in many cell types including RPE but not in melanocyte-lineage cells." - http://www.uniprot.org/uniprot/O75030
Nina: Thus, we are interested in the isoform M of MITF.
“MITF Human Recombinant (aa 170-279) expressed in E.coli, shows a 38 kDa band on SDS-PAGE.” - http://mbl.se/ViewProduct.aspx?id=148088
“There are two known isoforms of MITF differing by 66 amino acids at the NH2 terminus. Shorter forms are expressed in melanocytes and run as two bands at 52kDa and 56kDa, while the longer MITF form runs as a cluster of bands at 60-70kDa in osteoclasts and in B16 melonoma cells” - http://mbl.se/ViewProduct.aspx?id=148088
"“Wild-type His-MITF [...] were expressed in E.coli BL21 (DE3)”" - Ser298 of MITF a mutation site in Waardenburg syndrome type 2 is a phosphorylation site with functional significance
Nina: This article and the following three shows that other research groups have produced MITF already in bacteria such as e.coil (however as in a fusion protein, never alone), which is good for us, knowing this and also looking at their protocols.
“MITF or STAT3 GST fusion proteins were expressed in protease-deficient E.coli strain B12” - Identifying a common molecular mechanism for inhibition of MITF and STAT3 by PIAS3
“To produce purified MBP fusion proteins, MBP–Mitf was expressed in Escherichia coli BL21 (DE3) and cultured in an incubator at 37 °C. The expression of the MBP–Mitf fusion protein was induced by the addition of 1 mM IPTG” - Construction of Protein Chip to Detect Binding of Mitf Protein (Microphthalmia Transcription Factor) and E-box DNA
“We produced Mitf as a maltose-binding protein (MBP) fusion protein in Escherichia coli” - Construction of Protein Chip to Detect Binding of Mitf Protein (Microphthalmia Transcription Factor) and E-box DNA
Catalase
Johan: At least E. coli and Bacillus (but not Lactobacillus) are catalase-positive, so it shouldnt be too hard to get it expressed/folded in those
"Catalase is an enzyme that effectively scavenges the toxic reactive oxygen species. Low levels of catalase have been demonstrated in the epidermis of patients with vitiligo.[3] UV irradiation of the skin is known to trigger free radical generation and resultant melanocyte damage. Human melanocytes in cell culture are especially sensitive to exygen radicals and melanocytes from vitiliginous skin require the addition of catalase to the culture medium to grow. The melanocytes from healthy skin proliferate without addition of catalase enzyme. Histological examination of involved and uninvolved epidermis in vitiligo revealed evidence for peroxidative damage to both keratinocytes and melanocytes." - Efficacy of antioxidants as an adjunct to photochemotherapy in vitiligo: A case study of 30 patients
Nina: This article I found shows the importance of catalase in skin.
"it appears that both involved and uninvolved epidermis of patients with vitiligo show abnormalities in antioxidant defense enzyme levels primarily in the keratinocyte, indicating that vitiligo could be a disorder of keratinocytes."- Efficacy of antioxidants as an adjunct to photochemotherapy in vitiligo: A case study of 30 patients
Nina: Thus both the keratinocytes and melanocytes are abnormal in vitiligo skin
"Escherichia coli produces two catalases or hydroperoxidases, HPI and HPII. HPI is a bifunctional catalase-peroxidase active as a tetramer of identical 80 049-Da subunits [...] HPII is a monofunctional catalase active as a tetramer of identical 84 118-Da subunits" - Probing the structure of catalase HPII of Escherichia coli — a review (1999)
Johan: I wonder if the monomers are stable, or if the entire tetramer has to be sent through the skin. I doubt a molecule with the mass of 320 kDa can go through..
Nina: Yes this is true what your saying, that would be too big..We have to check this out more.
"LACTOBACILLI are considered to be catalase-negative1, or occasionally to produce very small amounts of catalase2. The lactobacilli isolated from dairy products have always been found to be catalase-negative3,4. During recent work on the microflora of Cheddar cheese, we have encountered strains of lactobacilli which possess both strong and weak catalase activity. Three strains of Lactobacillus plantarum, originally isolated by Sherwood3 from New Zealand Cheddar cheese, were found to be catalase-positive. Two of the strains gave strong catalase reactions whereas the other strain was much weaker (see Table 1)." - http://www.nature.com/nature/journal/v178/n4535/abs/178700a0.html
Nina: This information is good, since we thought that there would not be any chance lactobacillus would express catalase.
"Even though our activated engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough H2O2 to be cytotoxic and then fissle, killing only themselves and little else. To avoid this problem, an E. coli catalase will be expressed by default, and then turned off shortly after pyruvate oxidase expression is triggered. We are accomplishing this regulation by putting katG (on of two E. coli catalase genes) behind the TetR sensitive promoter (pTet) and having TetR co-transciptionally expressed with the oxidase. The time it takes for TetR to accumulate in the cell provides the delay in repressing KatG expression. To ensure KatG is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted. The final strain will have deletions of both catalases, ensuring that we have complete control over catalase expression. " - https://2008.igem.org/Team:Caltech/Project/Oxidative_Burst
Nina: an iGEM team that has already expressed ctalase in e.coli, maybe we can get their gene from hq iGEM.
Superoxide dismutase
“[...] are a class of enzymes that catalyze the dismutation of superoxide into oxygen and hydrogen peroxide. As such, they are an important antioxidant defense in nearly all cells exposed to oxygen. One of the exceedingly rare exceptions is Lactobacillus plantarum and related lactobacilli, which use a different mechanism.” - http://en.wikipedia.org/wiki/Superoxide_dismutase
"Histological examination of involved and uninvolved epidermis in vitiligo revealed evidence for peroxidative damage to both keratinocytes and melanocytes. Superoxide dismutase is important for the generation and disproportionation of superoxide anion. As a consequence of UV irradiation, superoxide anion and reactive oxygen species are generated in the skin." - Efficacy of antioxidants as an adjunct to photochemotherapy in vitiligo: A case study of 30 patients
Nina: Superoxide dismutase removes accumulation of superoxide anion
"To offset the harmful effects of ROS, cells have evolved protective mechanisms that utilize antioxidant enzymes such as superoxide dismutases (SODs) and hydroperoxidases, which scavenge superoxide radicals and hydrogen peroxide" - Expression of a Heterologous Manganese Superoxide Dismutase Gene in Intestinal Lactobacilli Provides Protection against Hydrogen Peroxide Toxicity
Nina: There seems to be several SODs. We have to check out which one is preferable for us.
"Superoxide dismutases (SODs) (EC 1.15.1.1) are metalloenzymes that catalyze the conversion of the superoxide anion into hydrogen peroxide and dioxygen (38). There are three forms of the enzyme that are distinguished by their metal center: manganese, copper-zinc, or iron (25). These enzymes are found across a broad range of organisms, which can use one, two, or all three enzymes to meet their antioxidant needs (25). For example, Escherichia coli possesses all three isoforms (7, 32, 60). In most Streptococcus and Lactococcus spp., elimination of ROS conform to this general antioxidant defense system since they both possess MnSOD (46, 52). Previously, our group has identified, characterized, and cloned the gene (sodA) encoding the manganese-containing SOD from Streptococcus thermophilus AO54 (1, 12). Unlike most sodA genes, S. thermophilus sodA is expressed under both anaerobic and aerobic conditions and is not induced by the redox cycling compound, paraquat (12). This antioxidant enzyme was shown to be essential for the growth of S. thermophilus under aerobic conditions (1). However, most lactobacilli lack this general defense system (SODs). Lactobacillus plantarum developed an alternative nonenzymatic defense system that involves the accumulation of high intracellular concentrations of manganese ions, which can scavenge O2 ·� (2). The lack of endogenous SODs (49) and catalase may account for the high sensitivity of most species of Lactobacillus to oxidative stress. In this study, we report the expression of the sodA from S. thermophilus in four lactobacilli: L. gasseri NCK334, L. johnsonii NCK89, L. acidophilus NCK56, and L. reuteri NCK932. Under the conditions used in this study, the enzyme provided a clear protection against the inhibitory effect of H2O2 on the growth of L. gasseri and L. acidophilus." - Expression of a Heterologous Manganese Superoxide Dismutase Gene in Intestinal Lactobacilli Provides Protection against Hydrogen Peroxide Toxicity
"Recombinant Human Superoxide Dismutase produced in E. coli. is a stable dimer of two identical subunits, non-glycosylated, containing 308 amino acid residues, two pairs of disulfide bonds and having a combined molecular mass of 31.6kD." - http://www.gentaur.com/recombinant_human_sod.htm
Nina: here we see that a company has produced Human Superoxide Dismutase in e.coli. We might also do this, instead of introducing non human (bacterial) Superoxide Dismutase.
Mast cell growth factor (MGF)
Nina: This one is preferred to introduce in vitiligo skin since it stimulates melanocytes by hair follicles to migrate up to their final position in the skin, which is the basal layer of epidermis.
Basic fibroblast growth factor (bFGF)
“Vitiligo is a skin disease that is caused by selective destruction of melanocytes and is characterized by white spots. Melanocytes and keratinocytes seem to exhibit a functional close relationship, mediated at least in part by keratinocyte-derived cytokines, which seem important for survival and activity of melanocytic cells” - New Insights into the Pathogenesis of Vitiligo: Imbalance of Epidermal Cytokines at Sites of Lesions p. 87
Nina: These cytokines are important for us to study in order to check if we can introduce any into the skin.
"It has been hypothesized that melanocytes form a functional unit together with keratinocytes, the so-called epidermo-melanic unit (6). Close relationships important for melanocyte survival and differentiation likely exist between these two cell types, that are mainly because of keratinocyte-derived cytokines, acting on melanocytes via speci®c receptors (7, 8). Keratinocytes secrete GM-CSF, an intrinsic stimulant of melanocytes in UVA-induced melanosis (9), endothelins, which are intrinsic mitogens for melanocytes and stimulate UVB hyperpigmentation (10), and bFGF which is a natural mitogen for melano-cytes (11) and promotes melanin synthesis (12). SCF/c-KIT pathway mediated by keratinocytes plays a critical role in the control of normal human melanocyte home-ostasis, increasing the number, size, and dendricity of melanocytes (13). Furthermore, keratinocytes synthesize IL-1a, IL-6, TNF-a and TGF-b, which are paracrine inhibitors of human melanocyte proliferation and mela-nogenesis (14±18). Therefore, it is possible to hypothesize that a functional change of cutaneous microenvironment ± i.e. an impairment in keratinocyte secretory activity ± may be involved in melanocyte disappearance in vitiligo." - New Insights into the Pathogenesis of Vitiligo: Imbalance of Epidermal Cytokines at Sites of Lesions p. 88
Nina: The bFGF is interesting for us to continue studying. It does not need any glycolisation, which GM-CSF does. SCF stand for stem cell factor and might induce all types of cells to proliferate and get to active which might result in cancer development. However bFGF might also stimule for this, but probably not in the same extent, making it better for us to work with.
Nina: There are already antibodies against the cytokines, which is good since we don’t need to produces them in this case:
Anti-SCF
Anti-bFGF --> Good
Anti-GM-CSF -non usable since need glycolisation
Anti-sense since these are overexpressed and we want to have them downregulated:
Anti-IL-6
Anti-TNF-a
Anti-TGF-b1±2-3
Anti-GM-CSFR
Anti-TNFR
Anti-IL-6R
-New Insights into the Pathogenesis of Vitiligo: Imbalance of Epidermal Cytokines at Sites of Lesions p. 88
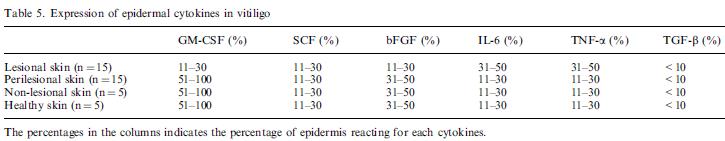
Nina: is a table showing Expression of epidermal cytokines in vitiligo skin from biopsis. The percentages in the columns indicates the percentage of epidermis reacting for each cytokines.
Nina: Mats also thought was good of us to pick bFGF for studying
“Research on mechanisms by which melanocytes migrate from the lower hair follicle to the epidermis has focused on cytokine release by keratinocytes, i.e. leukotriene C4 (LTC4), endothelin-1 (ET-1), basic fibroblast growth factor (bFGF), and stem cell factor (SCF), all of which stimulate melanocyte migration.46 As such, it was found that LTC4 and transforming growth factor a (TGF-a), released from keratinocytes secondarily to UV damage, stimulate melanocyte migration in vitro.47 Attachment of cells to the extracellular matrix and their migration over the matrix are mediated by the binding of domains of the matrix proteins with integrin receptor on the cell surface.48 Cultured melanocytes were shown to have an increased random migration on type IV collagen and to express cell membrane a2b1, a3b1, and a5b1 integrins,49 which were shown to be important in controlling the migration of many cell types; similar roles were attributed to the component proteins of focal contacts such as vinculin, talin, a-actinin, and paxilin, which play a critical role in directed cell migration, adhesion, and normal growth." - New Insights onTherapy with Vitamin D p 517
“Recombinant Human FGF-basic (FGF-2) produced in E.Coli is a single, non-glycosylated, polypeptide chain containing 155 amino acids and having a molecular mass of 17353 Dalton” - http://www.imgenex.com/recomb_tds.php?id=1036
Nina: This article and the following are good since they says that bFGF has been produced in E.coil, which we also want to do.
“In summary, we have expressed and purified a GSTbFGF fusion protein from E. coli. The fusion protein can stimulate the growth of HUVEC, indicating that GST does not interfere with bFGFs activity.” - Expression and purification of a biologically active basic fibroblast growth factor fusion protein p. 270
"[...] bFGF is an 18 kDa protein with a length of 155 amino acids and an isoelectric point of 9.6." - http://www.copewithcytokines.de/cope.cgi?key=bFGF
Nina: I found out that there were many of these bFGFs, so I searched and found that the smaller one, which is the 18 kDa is the one of interest for us, but I need more information about this.
"[...] occurs predominantly in the cytosol" - http://www.copewithcytokines.de/cope.cgi?key=bFGF
"Our results showing that 18-kDa FGF-2 is predominantly localized to the nucleus are consistent with most but not all (7–16) studies of subcellular distribution of 18-kDa FGF-2. Some reports show that 18-kDa FGF-2 is predominantly cytosolic (17–20). The reason for the discrepancy is not clear." - Nuclear and Nucleolar Localization of 18-kDa Fibroblast Growth Factor-2 Is Controlled by C-terminal Signals p. 40159
Nina: bFGF 18 kDa is located in both the cytoplasm & nucleus.
Vitamin D
Nina: Vitamin D has shown to be beneficial for vitiligo patients. Vitamin D would be secreted from the bacteria, penetrating the skin, and enter the melanocytes.
“A concept regarding the implication of non-genetic factors in vitiligo etiopathogenesis is supported by morphological and functional evidence that vitiligo is a disease of the entire epidermis with biochemical abnormalities within keratinocytes that influence the ability of melanocytes to counteract the reactive oxygen species (ROS); the morphological alterations of keratinocytes in the skin were believed to alter subsequently the production of the specific melanocyte growth factors and cytokines that coordinate melanocyte activity.17 The in- creased levels of hydrogen peroxide found in affected epidermis are supposed to lead to membrane damage.17 Moreover, oxidative stress may lead to hypopigmentation by me- chanisms that include a microphthalmia-associated transcription factor (MITF)-dependent downregulation of melanogenic enzymes; this is in good agreement with the deficiency of MITF-M described in vitiliginous skin.31 In addition, cultured keratinocytes and melano- cytes from depigmented vitiligo epidermis have been shown to have a reduced enzymatic antioxidant capacity,17,32,33 thus making the melanocytes abnormally susceptible to im- munologic cytotoxicity and cytotoxicity induced by ROS.27 A growing consensus supports the hypothesis that genetic factors and environmental triggers lead eventually to autoimmune melanocyte destruction.2,14 The altered environment of vitiligo skin, apparently rich in ROS17,34 and having an imbalanced cytokine network, may also contribute to additional melanocyte cytotoxicity and further enhance the pigment loss.“ - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo
“Furthermore, vitamin D and its chemically engineered analogs are supposed of being involved in the control of multiple intracellular pathways responsible for the melanin synthesis and melanocyte survival and in the suppression of the immune response. All these findings encouraged clinicians to further investigate the effects of treatment with vitamin D compounds in vitiligo patients, thus opening new perspectives in the therapy of depigmentation.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, pp. 18-19
Nina: This seems to be a good vitamin to take into vitiligo skin, however if would be great if we could find any article about vitamin d production in bacteria such as e.coli or lactobacillus.
“Vitamin D ligands control multiple critical biological functions and play different roles depending on the type of the tissue targeted and on the mechanism(s) involved in the cellular signaling. These compounds, acting mainly through their nuclear receptors (vitamin D receptor—VDR), protect the epidermal melanin unit and restore the melanocyte integrity by two main mechanisms (reviewed in Birlea et al.33): controlling the activation, proliferation, and migration of melanocytes and the pigmentation pathways, and modulating the T cell activation that is apparently correlated with the melanocyte disappearance in vitiligo. The multiple effects of VDR on immune cells lead to the recognition of vitamin D as a potent immunomodulator.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.19
“Experiments using normal melanocytes62,63 and melanoma cells64 of animal and human origin showed a stimulatory effect of vitamin D compounds on melanocyte differentiation and tyrosinase activity. Studies showed that topical application of vitamin D3 on the epidermis of mice increased the number of DOPA-positive melanocytes.65 Indirect data about the stimulatory effect of vitamin D on tyrosinase were provided by animal models fed with vitamin D, which in contrast with vitamin D-deficient animals showed a greater skin tyrosinase activity in response to UV irradiation.66” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.19
(“Opposed to its stimulatory effects on melanocyte differentiation, vitamin D was also reported to have an inhibitory effect on the melanocyte growth,67–69 an effect that was confirmed on several other cell types as well. As such, vitamin D has been shown to act as a regulator of replication in keratinocytes, and has been reported to coordinate the growth arrest in several types of cancer cells such as prostatic, breast, colon, leukemic, squamous carcinoma, melanoma, and Kaposi sarcoma cells.56,70,71 It may be inferred that while the stimulatory effect of vitamin D compounds on tyrosinase activity and the modulatory effect on T cell infiltrate may enhance repigmentation in vitiligo, their suppressor effect on melano- cyte growth would minimize or delay the pigment restoration. Therefore, designing new vitamin D analogs able to stimulate melanocyte differentiation and to enhance tyrosinase activity is of a particularly high interest in vitiligo.
The mechanism through which vitamin D exerts its effects on melanocytes is not yet fully understood. It is believed that vitamin D is involved in melanocyte physiology by coordinating the melanogenic cytokines (most likely endothelin-3 (ET-3)) and the activity of SCF/c-Kit system—one of the most important regulators of melanocyte viability and maturation.33” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.20)
“Furthermore, a proposed mechanism involving vitamin D in the protection of vitiliginous skin is based on its antioxidant properties and regulatory function toward the ROS that are produced in excess in vitiligo epidermis. It is also believed that vitamin D coordinates calcium fluxes in melanocytes, a similar function to that exerted in other cell types such as human keratinocytes, hepatocytes, and murine B16 melanoma cells.33,72,73 Based on these observations, it has been suggested that vitamin D is involved in the correction of the aberrant calcium fluxes that accompany the melanocytic loss in vitiligo.33,34,74,75 Several other studies emphasized the complex anti-apoptotic action of vitamin D on melanocytes76 and keratinocytes.77,78 It is likely that vitamin D can protect pigment cells against apoptosis, one of the cytotoxic mechanisms proposed among those causing melanocyte disappearance in vitiligo.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.20
Nina: We need to apply the vitamin d into our bacteria that has been documented having the shortes pathway.
Andreas: Keep in mind that vitamin D (in the human body at least) is produced from a precursor that requires (UV?) light to be converted into vitamin D. Maybe this does not apply to bacteria, I don’t know.
Nina: Size of vit D: ~400 g/mol
Vitamin B12 (cyanoocobalamin)
“The biosynthesis pathway has been extensively characterized in several lactic acid bacteria, including Lactobacillus plantarum WCFS1” - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
Nina: This is good, allowing us to work in lactobacillus with this vitamin.
"The constitutive overexpression of the folate biosynthesis genes of L. plantarum WCFS1 in cultures of L. reuteri JCM1112/pNZ7026 resulted in an almost 100-fold increase in folate levels, while the control (L.reuteri JCM1112/pNZ7021) did not show any change in folate and B12 production. The overproduction of folate was found to have a very small effect on B12 production (10% reduction), resulting in a folate/B12 ratio of approximately 100:1 (wt/wt)" - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
Nina: This means and the following two citations show that we can have our lactobacillus producing both vitamin B without disturbing each others pathways.
“The same construct has been tested with L. plantarum, resulting in similar folate production levels (28), and similar results were obtained when the same strategy was applied to Lactococcus lactis (30) and Lactobacillus gasseri (29).” - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
“In this study, we demonstrated that it is possible to combine the production of folate and the production of B12 in L. reute” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Vitamin B12 consists of a tetrapyrrolic-derived corrin ring with a cobalt ion chelated at the core” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Coenzyme B12 biosynthesis is limited to a few representatives of bacteria and archaea (Martens et al., 2002). It appears that B12-dependent enzymes are absent from plants and fungi, but widespread in prokaryotes, protists and animals” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Lb. reuteri was the first lactic acid bacterium reported to be able to produce B12” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
Nina: This means there should be many more articles about Lb. reuteri and vitamin B12. We need to learn more.
“In this study, we extend the analysis of the presumed coenzyme B12 biosynthesis gene cluster of Lb. reuteri and describe the presence of a complete gene cluster encoding all the enzymic machinery necessary for the de novo synthesis of this important cofactor.” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
Nina: Size of vit B: ~440/1300 g/mol
Vitamin B9 (folic acid)
"Lactobacillus plantarum WCFS1 wild-type and L. plantarum engineered strains harboring pNZ7021 (control vector, (41)) and pNZ7026 (folate gene cluster expression vector (40" - Modulation of Folate Production in Lactic Acid Bacteria p. 131
Nina: This plasmid (pNZ7026 ) is very good for us to have in the project. They have expressed vitamin B9 in Lactobacillus plantarum.
"Folate was quantified using a microbiological assay on samples which were pretreated with γ-glutamyl carboxypeptidase (E.C.3.4.19.9) for deconjugation of polyglutamate tails (17, 33). A qualitative folate assay was developed and used for high throughput analysis of the samples." - Modulation of Folate Production in Lactic Acid Bacteria p. 131
Nina: Maybe we also could use this assay to quantify our vitamin B9 production in our bacteria.
Andreas: The 2008 Caltech iGEM team had a subproject involving folate/vitamin B9 production in E. coli. They cloned the fol genes, and the pab genes from Lactococcus lactis in E. coli. Some of the genes have been submitted to the Registry. https://2008.igem.org/Team:Caltech/Project/Vitamins
Secretion
"A number of methods have been applied to promote extracellular secretion of recombinant proteins from E. coli. These include the use of biochemicals, physical methods (osmotic shock, freezing and thawing), lysozyme treatment, and chloroform shock. However, these methods can be applied only after harvesting cells" - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, p.630
"E. coli normally does not secrete proteins extracellularly except for a few classes of proteins such as toxins and hemolysin. Secreted proteins can leak from the periplasmic space into the culture medium possibly due to an increased permeability of the cell membrane during a lengthy incubation period. Small proteins secreted into the periplasm are frequently released into the culture medium (Tong et al. 2000). In general, movement of recombinant proteins from the periplasm to the culture medium is the result of compromising the integrity of the outer membrane. However, care must be exercised during such recombinant protein production so as not to compromise cellular integrity, which often causes cell death" - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, pp.630,632
Nina: It is good that e.coli does not secrete many types of proteins since we then would have this bacteria secreting somewhat only our proteins and other molecules of interest.
"Another method of extracellular protein production involves fusing the product to a carrier protein that is normally secreted into the medium (e.g. hemolysin), or to a protein expressed on the outer membrane (e.g. OmpF). For example, human β-endorphin could be secreted into the culture medium when fused to OmpF (Jeong and Lee 2002; Nagahari et al. 1985). Recently, a method of releasing active scFv antibody and human interleukin-6 into the culture medium using the hemolysin secretion pathway was reported (Fernandez et al. 2000; Li et al. 2002). The hemolysin transport system (Hly) is a type-I secretory apparatus that forms a protein channel between the inner and outer membranes o fE. coli. Hemolysin toxin (HlyA) is secreted by direct passage of the HlyA polypeptide from the cytoplasm to the extracellular medium using the hemolysin transport system (Fig. 2). For extracellular production using the hemolysin secretion pathway, the target protein is fused to the C-terminal hemolysin secretion signal. The Hly system appears to be an attractive candidate for the extracellular production of recombinant proteins." - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, p.632
"Proteins secreted into the E. coli periplasm can also be released into the culture medium by co-expression of kil (Kleist et al. 2003; Miksch et al. 1997; 2002; Robbens et al. 1995) or the gene coding for the third topological domain of the transmembrane protein TolA (TolAIII) (Wan and Baneyx 1998). Zhou et al. (1999) reported extracellular production of the Erwinia chrysanthemien- doglucanase by employing the out genes from E. chrysanthemi EC16, which are responsible for the efficient extracellular secretion of pectic enzymes. Co-expression of the out genes increased production of active endoglu- canase and released enzymes equivalent to over half of the total activity into the extracellular medium." - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, p.632
"Another approach to the extracellular production of target proteins uses L-form cells, wall-less, or wall- deficient cells (Kujau et al. 1998; Rippmann et al. 1998). Recently, Kujau et al. (1998) demonstrated that L-form E. coli cells were capable of secreting into the culture medium a recombinant antibody fragment (single- chain phosphorylcholine-binding scFv from human McPC603) fused to the OmpA signal sequence under the control of the lac promoter. A correctly folded and dimerized mini-antibody was secreted directly, and remained stable in the culture medium." - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, p.632
"Bacteriocin release protein (BRP) can also be used in the extracellular production of recombinant proteins in E. coli. BRP is a 28-amino-acid lipoprotein that activates detergent-resistant phospholipase A, resulting in the formation of permeable zones in the cell envelope through which proteins can pass into the culture medium (Fu et al. 2003; Lin et al. 2001b; van der Wal et al. 1995). However, co-expression of the BRP gene can damage the cell envelope and cause release of other cellular proteins. Recently, van der Wal et al. (1998) reported that a modified BRP gene (Lpp-BRP) could be used for the extracellular production of K88 fimbrial molecular chaperone FaeE without growth inhibition, lysis, or contaminating proteins." - Secretory and extracellular production of recombinant proteins using Escherichia coli, 2004, p.632
Johan: Maybe there has been some tricks developed the last 5 years that we can take advantage of (anyone?), otherwise it seems that we have to use http://partsregistry.org/wiki/index.php?title=Part:BBa_J32015 and maybe get it to leak to the outside? (anyone disagree?). For Lactobacillus it seems to be ezmode, just add http://partsregistry.org/wiki/index.php?title=Part:BBa_K128004
Choise of chassis
"In iGEM, and synthetic biology in general, the microorganism that is being worked with is referred to as the cellular chassis (shăs'ē, chăs'ē). For most iGEM teams, the chassis will be E. coli, but you are not limited to working with E. coli in iGEM. Some of the past chassis have been Bacillus subtilis, lactobacillus, yeast, and even some mammalian cells.
So, why does it matter that different chassis are used, anyway? Well, beyond their different growth requirements and quirks about competency (the ability of an organism to take up DNA), different chassis have different innate abilities. These innate abilities may make the chassis more worthwhile to work with than another microbe, e.g., E. coli. Because while E. coli is the best researched microbe, it may simply not have a trait you need for your project." - http://umassigem.blogspot.com/2009/02/concepts-cellular-chassis.html”
"We have observed that proteins, such as human tissue-type plasminogen activator, pro-urokinase or gp41 of human immunodeliciency virus, which have a high content of rare codons in their respective genes, are not readily expressed in Escherichia coli. Furthermore induction of these heterologous genes leads to growth inhibition and plasmid instability. Supplementation with tRNAhAaGA,AGGby cotransfection with the dnaY gene, which supplies this minor tRNA, resulted in high-level production with greatly improved cell viability and plasmid stability." - High-level expression of recombinant genes in Eschericlriu coli is dependent on the availability of the dnaY gene product
Johan: Interesting way of increasing expression by adding a gene for a rare tRNA
"Within Escherichia coil and other species, a clear codon bias exists among the 61 amino acid codons found within the population of mRNA molecules, and the level of cognate tRNA appears directly proportional to the frequency of codon usage. Given this situation, one would predict translational problems with an abundant mRNA species containing an excess of rare low tRNA codons. Such a situation might arise after the initiation of transcription of a cloned heterologous gene in the E. coli host. Recent studies suggest clusters of AGG/AGA, CUA, AUA, CGA or CCC codons can reduce both the quantity and quality of the synthesized protein. In addition, it is likely that an excess of any of these codons, even without clusters, could create translational problems." - Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli
Johan: Example of article about codon bias, although if e.g. we’re gonna do DNA synthesis at GENEART (50% off) it is done automatically (what about their “The minimum charge for each DNA synthesis projects is $159” ???)
- Foreign Gene Expression in Yeast: a Review
Johan: Has many factors for yeast, some things can be taken into consideration for bacteria too
• E. coli (Super easy to work with in the lab)
• Bacillus subtilis (The model organism of gram positives, of which Lactobacillus also is a member of)
• Lactobacillus (Very friendly for the skin)
* https://2007.igem.org/Edinburgh/Yoghurt/Design#Gene_Expression_In_Lactobacillus * https://2008.igem.org/Team:MIT#Lactobacillus_Work * http://openwetware.org/wiki/Category:Lactobacillus * http://openwetware.org/wiki/Electrotransformation_of_Lactobacillus_Spp. * http://openwetware.org/wiki/Cfrench:BioBrickVectors1#Making_pTG262_into_a_BioBrick_vector
"“Our decision to use L. plantarum in our experiments was based on the fact that there are no reports indicating the possible virulence activity of L. plantarum in experimental models (15) or in spontaneous infections (14). Although lactobacilli have an excellent overall safety record among probiotics, there are a few reported cases of infection in premature neonates with severe immune deficiencies caused mainly by Lactoba- cillus rhamnosus (21).”" - Bacteriotherapy with Lactobacillus plantarum in burns, p.79
Andreas: Found some information on using Lactobacilli as our cloning organism for the project. The Edinburgh 2007 iGEM team used a Lactobacillus strain (probably acidophilus?) for their yoghurt project. They created a vector which they successfully used in both Lactobacillus and E. coli. The vector is available as BBa_I742103 and the project link is attached below: https://2007.igem.org/Edinburgh/Yoghurt/Design#Gene_Expression_In_Lactobacillus I also found out that many Lactobacilli have pretty short generation times (down to 25 minutes), so that shouldn’t be a problem either.
Andreas: They have now released the contents of the DNA kit plates which will be sent out to the teams. BBa_I742103 is not included in these kits, which means we will have to order it separately. It might be a good idea to check whether there are any other BioBricks we need that are not included in the kits, so that we can order them from the BioBrick foundation. Such parts might be antibiotic resistance cassettes, for example, if we need to use any specific antibiotics when working with Lactobacillus. Here’s a link to the DNA kits of the 2010 competition:
http://partsregistry.org/assembly/libraries.cgi?id=31
Proteins that have already been expressed
- Cell penetrating peptide (CPP) - Yes (e.coli)
...
- Protein A - Yes (e.coli)
...
- IgG protease Yes (Stafylocoous)
...
- Melanin -Yes (e.coli)
...
- MITF (Microphthalmia-associated transcription factor) - Yes (e.coli)
...
- Catalase - Yes (e.coli & lactobacillus)
...
- Superoxide dismutase - Yes (from Streptococcus thermophilus AO54 then in lactobacillus)
...
- Mast cell growth factor (MGF)
...
- Basic fibroblast growth factor (bFGF) - Yes (e.coli)
...
- Vitamin D
...
- Vitamin B12 (cyanoocobalamin) - Yes (lactobacillus)
...
- Vitamin B9 (folic acid) - Yes (lactobacillus)
...
Practical schedule
Nina: I am putting this part together since we need some directions for the following months.
Team members
- Nina Schiller - Johan Nordholm - Andreas Constantinou - Emmelie Lidh - Hassan Asl Foroughi
Time plan
Theoretical studies: week ~6 Practical starts: week 21-22 Practical ends: week ~41 Theoretical studies: week ~42-44
Wet lab
May/June
- Constructing biobricks - insert gene of interest into iGEM vectors
- Sequence the constructs (to check if they are corrects)
- Cell culturing both bacteria and eukaryotic cells
- Save one version of biobricks in freezer to send in and transform other version into bacteria
July
- Study expression / characterizing molecule
- Study secretion / characterizing molecule
August
- Study expression / characterizing molecule
- Study secretion / characterizing molecule
September
- Study expression / characterizing molecule
- Study secretion / characterizing molecule
- Study passage of molecules through skin
October
- Study passage of molecules through skin
- Send in the one version of biobricks saved in freezer
Wet-lab
- Nina: CPP, Protein A, Tyrosinase, IgG protease
- Johan: CPP, bFGF
- Andreas: CPP, SOD+yCSS
- Emmelie: CPP, MITF
Jamboree
November week 44-45
Subcellular locations of molecules in eukaryotic target cell
To see if they are compatible with CPP (e.g. will probably not insert to a membrane).
- Protein A - outside of eukaryotic target cell
- IgG protease - outside of eukaryotic target cell
- Vitamin D - nucleus
- Vitamin B9
- Vitamin B12
- Melanin - cytoplasm
- Catalase - Peroxisome (...)
- Superoxidase dismutase - 3 forms. One in the cytoplasm, one in the mitochondrion matrix and one extracellular
- bFGF - cytoplasm & nucleus
- MFG
- MITF - nucleus
Post-translational modifications
Must be taken into account for prokatyotic expression, dependent on how vital the modifications are.
- Protein A -
- IgG protease ?
- Vitamin D -
- Vitamin B9 -
- Vitamin B12 -
- Melanin -
- Catalase - "The N-terminus is blocked"
- Superoxidase dismutase -
1st: "The displayed sequence is further processed into a mature form.", "Unlike wild-type protein, the pathogenics variants ALS1 Arg-38, Arg-47, Arg-86 and Ala-94 are polyubiquitinated by RNF19A; which leads to their proteasomal degradation."
2nd: "Nitrated under oxidative stress. Nitration coupled with oxidation inhibits the catalytic activity."
3rd: ?
- bFGF - "Several N-termini starting at positions 94, 125, 126, 132, 143 and 162 have been identified by direct sequencing"
- MFG: ?
- MITF - "Phosphorylation at Ser-405 significantly enhances the ability to bind the tyrosinase promoter"
Parts available at iGEM hq and elsewhere
- Protein A (Z domain) - Yes, a gift from prof. Mathias Uhlén at KTH
- IgG protease - Yes, a gift from prof. Ulrich von Pawel-Rammingen at Umeå University
- Vitamin D -
- Vitamin B9 - Yes, a gift from Dr. Eddy Smid at NIZO food research BV
- Vitamin B12 -
- Melanin - Yes, gene called melA from iGEM hq
- Catalase - Yes, gene called KatG from (Team:Caltech 2008) iGEM hq (BBa_K137079)
- Superoxidase dismutase - Yes, a gift from prof. Mikael Oliveberg at Stockholm University
- yCCS - Yes, a gift from prof. Mikael Oliveberg at Stockholm University
- bFGF - Yes, a gift from Dr. Nozomu Nishi at Kagawa University
- MFG -
- MITF - Yes, a gift from prof.Shigeki Shibahara at Tohoku University School of Medicin
- Promoter + RBS (ribosome binding site) - BBa_J04500 (IPTG+RBS), BBa_K215000 (strong IPTG+RBS), BBa_R0010 (Lacl+RBS) Both are compatible to assembly standard 25.
- Plasmid/plasmid backbone - Part:BBa_K243033 http://partsregistry.org/wiki/index.php/Part:BBa_K243033
http://partsregistry.org/wiki/index.php?title=Part:BBa_J63010 To get silver standard and hence in-frame fusions
- Resistance -
- Transcription termini -
- Secretion signal - http://partsregistry.org/wiki/index.php?title=Part:BBa_J32015 (already in our plates from iGEM!) (not compatible with standard 25 ...)
- His-tag - http://partsregistry.org/wiki/index.php?title=Part:BBa_K157011 (compatible with assembly standard 25)
- Linker (if needed) - BBa_J18920 2aa GS linker, BBa_J18921 6aa [GS]x linker, BBa_J18922 10aa [GS]x linker (all compatible with assembly standard 25)
- GFP - Part:BBa_E0040 http://partsregistry.org/Part:BBa_E0040
DNA sequences
What to look out for
Restriction sites
- BBa_M31982 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31982, NotI restriction enzyme site, gcggccgc
- BBa_M31983 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31983, EcoRI restriction enzyme site, gaattc
- BBa_M31991 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31991, XbaI restriction enzyme site, tctaga
- BBa_M31992 http://partsregistry.org/wiki/index.php?title=Part:BBa_M31992, SpeI restriction enzyme site, actagt
- PstI restriction enzyme site, ctgcag
Johan: Added PstI, why is not in the list on http://partsregistry.org/DNA/Assembly?
Search on e.g. http://mobyle.pasteur.fr/cgi-bin/portal.py?form=restrict
Andreas: Here are directions for constructing new parts:
http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication#Constructing_a_BioBrick_part_via_direct_synthesis
Restriction sites that must be targeted for removal:
- EcoRI (GAATTC)
- XbaI (TCTAGA)
- NotI (GCGGCCGC)
- SpeI (ACTAGT)
- PstI (CTGCAG)
- NgoMIV (GCCGGC) (Freiburg standard)
- AgeI (ACCGGT) (Freiburg standard)
Restriction sites that should be targeted for removal:
- ApoI (RAATTY) (probably impossible because it occurs frequently)
- MfeI (CAATTG)
- AvrII (CCTAGG)
- NheI (GCTAGC)
- NsiI (ATGCAT)
- SbfI (CCTGCAGG)
For details on why, check the article "Engineering BioBrick vectors from BioBrick parts", table 1.
CPP
Transportan 10 (Ülo Langel's)
Amino acid sequence:
AGYLLGKINLKALAALAKKIL-amide
- Mechanism of the Cell-Penetrating Peptide Transportan 10 Permeation of Lipid Bilayers
DNA sequence:
GCGGGTTACCTGCTGGGTAAAATCAACCTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTG
- (http://genomes.urv.es/OPTIMIZER)
Restriction sites: none
Low-molecular-weight protamine (LMWP) peptide
Amino acid sequence:
VSRRRRRRGGRRRR
- Synthetic Skin-Permeable Proteins Enabling Needleless Immunization
DNA sequence:
GTTTCTCGTCGTCGTCGTCGTCGTGGTGGTCGTCGTCGTCGT
- (http://genomes.urv.es/OPTIMIZER)
Restriction sites: none
TAT ("Dowdy's CPP")
Amino acid sequence:
YGRKKRRQRRR
- TAT-Mediated Protein Transduction into Mammalian Cells
Nucleotide sequence:
TACGGTCGTAAAAAACGTCGTCAGCGTCGTCGT
- (http://genomes.urv.es/OPTIMIZER)
Restriction sites: none
Protein A (Z domain)
GTAGACAACAAATTCAACAAAGAACAACAAAACGCGTTCTATGAGATCTTACATTTACCT
AACTTAAACGAAGAACAACGAAACGCCTTCATCCAAAGTTTAAAAGATGACCCAAGCCAA
AGCGCTAACCTTTTAGCAGAAGCTAAAAAGCTAAATGATGCTCAGGCGCCGAAA
- A synthetic IgG-binding domain based on staphylococcal protein A, p. 107
Restriction sites: ApoI (10)
IgG protease
ATGGATAGTTTTTCTGCTAATCAAGAGATTAGATATTCGGAAGTAACACCTTATCACGTT
ACTTCCGTTTGGACCAAAGGAGTTACTCCTCCAGCAAACTTCACTCAAGGTGAAGATGTT
TTTCACGCTCCTTATGTTGCTAACCAAGGATGGTATGATATTACCAAAACATTCAATGGA
AAAGACGATCTTCTTTGCGGGGCTGCCACAGCAGGGAATATGCTTCACTGGTGGTTCGAT
CAAAACAAAGACCAAATTAAACGTTATTTGGAAGAGCATCCAGAAAAGCAAAAAATAAAC
TTCAATGGCGAACAGATGTTTGACGTAAAAGAAGCTATCGACACTAAAAACCACCAGCTA
GATAGTAAATTATTTGAATATTTTAAAGAAAAAGCTTTCCCTTATCTATCTACTAAACAC
CTAGGAGTTTTCCCTGATCATGTAATTGATATGTTCATTAACGGCTACCGCCTTAGTCTA
ACTAACCACGGTCCAACGCCAGTAAAAGAAGGTAGTAAAGATCCCCGAGGTGGTATTTTT
GACGCCGTATTTACAAGAGGTGATCAAAGTAAGCTATTGACAAGTCGTCATGATTTTAAA
GAAAAAAATCTCAAAGAAATCAGTGATCTCATTAAGAAAGAGTTAACCGAAGGCAAGGCT
CTAGGCCTATCACACACCTACGCTAACGTACGCATCAACCATGTTATAAACCTGTGGGGA
GCTGACTTTGATTCTAACGGGAACCTTAAAGCTATTTATGTAACAGACTCTGATAGTAAT
GCATCTATTGGTATGAAGAAATACTTTGTTGGTGTTAATTCCGCTGGAAAAGTAGCTATT
TCTGCTAAAGAAATAAAAGAAGATAATATTGGTGCTCAAGTACTAGGGTTATTTACACTT
TCAACAGGGCAAGATAGTTGGAATCAGACCAATTAA
Restriction sites: AvrII (420), NsiI (779)
Contains 1 AvrII & 1 Nsil, not so important
Vitamin D
-
Vitamin B9
Vitamin B9 cluster
>pNZ7026 pCon + folate gene cluster van plantarum
CGGACTAATCATTATTCGATAACCAATAGCCATTCTCAATTCGGCCTAACAGCGTAATCA
ATTGCTTGGTTTCAGCCACATCGTGGACCCGGATGATCCGGCCACCCTTAAGCATCATCG
CTGCTTCAGTGACCAACGTCATTGGTAGCCGGTCTTCCTTCTTCAGACCAAGCAATAGAC
CTAAATAACCTTTCCGTGAAATGGCCACCATCATTGGGCGTCGCAAATAATTAAATTCAT
CAATGTTACGCATCATCACGTAATCCTGTTCCCCATGTGCGACTTGTGAATAGCCGATTC
CCTGATCTAGAGCAATTCGTTCAATATCGATGCCATGACTCGTCAATGCTGCCAGATTTT
GTTCAAAGAAACCACGCATACTGCTCGTCAGATCGGTATATTCTTGGTCACGACTACTGT
GCATAGTCAGTAAGCCGACGCGACTGTCAGCCATCAATTTGAGCTTGCGAGGGTCGTCCG
TAAAGGCGTTCACATCGTTAATAATGCTGACACCTTCTTCCAGGACGGCTTGCATGACAT
CATACTTATAAGTATCGACCGCTAAGACTGCTTCCGGGAAACGAGCCTTGATGGCGCGAA
TGTACGGCAACGTCCGGTCTAATTCAACTTGCGGCGTGACTTCCGTGAAGCCCGGCCGTG
TCGTTTGACCATTAACTTCGATTACGTCAGCTCCTGCTTGTAACATGGCATCCACGTGAT
TCAGTACGTCATCTTTAGTCTTGTACTGGCCACCGTCGAAAAAGGAGTCCGGCGTAATAT
TCATGATGCCATAAATCATCGGCTTCTTGGTGAGATTAAACCGGTGCTCACCCGCTTGCC
AAAAAACATCATATTGTTTCATAATTTGAGTCAATTGAATTTGAGTCGTTGCTTGATCCG
TAAAGACTCGCGACCATTGGTGTGTTAACTGGCGACCAGCTGTCTGGTTCAGCAAGACGG
TCAACTGTTCAGGACTCGCCTGCACGACCCCATCTAATTGCTGACATAGTTGGGTTAATC
GGAGCTGCTGCTCCCGATTATAATCACTAAAACGCAAAACTAATTGTTGTTGTCGTTGAA
CTTGGGCATTCAACGCCTGACTCGCAAAATCCGTTGCTTGTGCTAATGAACTGGTGATAT
CTTGTACTAACATCTACTCCACCGATCCTTCCAATTTTTAAAGTTTTGTGATTAATTTTA
AGGCGGCCCGTCCCCGGTGTGTCAGCGGGATGCGCCACGTTGCTGGTAATTCGGCCAAAG
TTCGACCATAGCGCGGTAGCCAGAAAATTCGGTCAAATCCCCCAGAATAGCGTCCAAATT
GATAGGACGCAATGTAGCCGGTGATCGAAGCTTGTACGGCCAAGATCCGCCCATCTGGCC
AGGCAGCCGCTAACGTAGCACGCATCAACGCTTGCCGCTCGTTGTCAGGGATATCGCGTA
GGGCCGTTAAAATTTCCTGATTGCAATCAAAGCCACTGACTGCAACCCCTAAGTCGCGCG
CCGTGGTCACACCTAACAAGTCTGGTAACGCGGAAATCTCTAAGCCACTATCATCTGCGA
TAACCGGAACCCCTAGCTGTTGCGCCCCAAAACGAGCTTTAGCAACCGCGTTATCGACAT
AGCTCGTCGTCGTTTCCACGGGAAATTCGAGCCGCGGCGCCACTGTCAAGTACTGACGAG
CAGTCAAGCCATAGTAAGCCAAACACGCGATCAAGTCGCGGCTTTTACCAGCGTTGTTGG
ATGCAATCAACCAAGTCGTCGTCATTGGCTTAACCCCGGCCAACCAGCCTGTTGGAGCTC
ACGAATGACATAAAATGACCCCGTTACAATAATCGCACTATTACTGGTTGCTTGCTGCTG
AGCTTGCGTTAACGCATCCGTGATCGTCGTTGCAGTCAATAAGTCTGGCTGTCGTTGCAT
CGCAGCCGTGGTCATAAAGGTGGGCGTTAATAATGCGTTCGCCGTGTCGGCCAGCTTAGT
CGCGGATAACGCTCGTTCTGGATTTGCGGGCGTATTCGTAATCAGGACGTCAGCTACCGG
TAATAGTTCAGCCAGCATTTTAGGGTAGGCCTTATCTTGTAAGACCCCGACGATCCAAAT
CAATTTACGACCAGCGAGTAATTGCTTCACACTTGCCACTAGTGCCGCCATACCATCCGG
ATTATGCGCCCCATCGGCAATAATGACAGGTTGCTCCTGTAATACGGTCAACCGGCCTGG
CAAGGAAACTTGTTGCAAGCCTCGGCGAACCGCACTGTTCGTCAAGTTAACTTGTTGCTG
TTGTAGTACTGCAACCACAGTTAAAACCAACCGCAGATTTTGTAGCTGATAGGTGCCACT
CAGTCCGACCGTTAATTGCGTCCAATCAAAAAGCTGACTGTGCGCATCAACGACGGTTTG
CGTGAGTGTTTGCCCAGTCACCGTCAGTTGCGCGTGCTTAGCGGTCACTAGCCGAACACC
TTGATTGAGCGCTTCGGTCTGCAAAACAGCCAGGGCTTCTGGGTGTTGGTCAGCCAATGT
CACTGCTGTTGTATGGGGCTTGATGATTTTTGACTTATTTTGCGCAATCGCGGTAATCGT
GGGCCCAAGAATGGCGGTATGATCGAGGGCAATCTTAGTAAATACCGTGAGTTGGGGACT
GGCTAAGGCGTTAGTCGCATCATACAAGCCGCCCAAACCGGCTTCAATGACCGCCCATTG
CACGTTTTGATTCCGAAACCAAACGACACTAATGAGAAAGAACCATTCAAAGATGGAGAC
ATCGCTAGCTGTCAGTCCCATATTTTGCAGAACCGGTAGAATCTCACGATACGTATCGAT
AAATTCCGCTGGACTGATCCACGTCCCATTGCATTGCAGTTGTTCACGCGCATCATTAAT
CGCGGGGCTGCTGAAGCGGCCAACTTGATACCCTTGCGCCCGCAGGATACTGGCTAACAT
GGCTCCCGTAGAGCCTTTGCCATTAGTCCCAGCGATATGGATCACATGGTAATAATGGTC
AGGGTGGCCAAGATGTGCCATGATGCGCCGTAATAACGGAACACGCTGATGATTATTGGC
TAACATGGCTTGATTTAGTTGGGCTAATAAGTCCTGATAACGTTGTTCAATCGTCCCTGT
ATCCACTTAATCACCTACTTTGCGATTCGCTGTAAGAATTCTCGTTTCAAGTCTAAATCC
GTCTTGAATTGACCACTGTAATAGCTACTTTCCGTATGCACACCCGGTTTGCTAACACCC
CGCATCTCCATGCACATGTGCCGCGCCGTGATTGAGACCGCGATCCCAGCCGGGTCAAGA
ATTCGTTGTAATTCTGTTGCAATGGAAACTGTCAACCGCTCCTGAACGTTCGGCTGTTGA
CTGCAATAGTCAATCAAGCGAGGAATCTTACTCAAGCCAATCACTTGTTCATGCTGCGGC
ACATAAGCAACTTGAACCGTGCCAAAAAACGGCAAAAGGTGATGCTCACACATCGAATAG
AATGGAATATCCTTAAGTAATACCATTTCAGTCGGATGCTCAACCTTGAACAGTTTATAA
TTATCAAATGGTGCGGCAGTCTTAGTAGCGAAAACTTCCGCATACATTCGTGCAACCCGT
GCTGGTGTTTCTACTAATCCCGGTCGGTCAGGATCTTCACCAACTGCACTTAAAATTTCT
CGTACTGCATGCTCAATCTTTGCTTGGTTCTTCTCATCAATCATCTAACTCACTCACATT
TCTTACTTTTTTAATCCAACTCTGATCCTGATTAGCCGCAATCAATTGGGCCACTTGTGG
TCCAACTAACACATCAGTTTTGGCGATTTCAGCAGTTGGCAGTAGCACAAAGTTGCGCTT
AGCTGCCTGCGCATGCGGAATCGTCAGCGTCGCTGTGTTGATTTGCCGGTCGCCCCAAAA
AATAATATCCAAATCAATCGTACGTGGTCCCCAGTGAACCAAGCGTTGGCGGTGGCCCGC
CTGCTCAATTGTATGTAATTCATCCAATAGTTCTTCTGGTGTCAAATTAGTCGTTAAGGA
TACCGAAACATTGTAAAAATTGGCCTGATCACGCTTTCCCCACGGTTGAGTCTCATACCA
ATGAGATTCGTCAATCACGTTTACCCCATTGACGGCTCGTAATCGGCTAAGGGCTTGCTG
AATATTTTGGACGCGCGGATGAATATTGGAACCAACACTCAAATAAACCCGTTCTTCCCT
ACTTGCCATTCGGCGTCCCCTCCACCTCAATTTCCACATCATCAAAGATTCCAGGCATTG
GTACGCTATATTTACGAATTTTAATATTGATTGCATCCGCCGCGGGAAAACTTGTCAATA
ACGTCTGCAATAAGTGGTTAGCTAGCGATTCAATCAACTTGTATGAATGGGTCGTTACAA
ATTCATCGACCACGTTACGGACCGCCGCGTAATTGATGGTCTCGTGAACGTCATCGTGTT
GAACCTTGGTTTCGATAGGATATTTAATGGCAATATCTAGCCCTAGTTGTTGACCATTAC
GCCGTTCTTCCGGAAGTACCCCGTTAAACGTGTGAAAGCGTAAATTATTAATTCGAATCA
TGCCCATGCCTTCTCCTAAATATTCAGTATTAATATAATTATATCAAATTGAATAGAAAA
TTTCGAGTACAAAGAAAAATTACATCTTTTCTTTAACTTATTAAAAAAACTGTCTACTGC
AGGTCGACAGATCTTGGAGCTGTAATATAAAAACCTTCTTCAACTAACGGGGCAGGTTAG
TGACATTAGAAAACCGACTGTAAAAAGTACAGTCGGCATTATCTCATATTATAAAAGCCA
GTCATTAGGCCTATCTGACAATTCCTGAATAGAGTTCATAAACAATCCTGCATGATAACC
ATCACAAACAGAATGATGTACCTGTAAAGATAGCGGTAAATATATTGAATTACCTTTATT
AATGAATTTTCCTGCTGTAATAATGGGTAGAAGGTAATTACTATTATTATTGATATTTAA
GTTAAACCCAGTAAATGAAGTCCAAGGAATAATAGAAAGAGAAAAAGCATTTTCAGGTAT
AGGTGTTTTGGGAAACAATTTCCCCGAACCATTATATTTCTCTACATCAGAAAGGTATAA
ATCATAAAACTCTTTGAAGTCATTCTTTACAGGAGTCCAAATACCAGAGAATGTTTTAGA
TACACCATCAAAAATTGTATAAAGTGGCTCTAACTTATCCCAATAACCTAACTCTCCGTC
GCTATTGTAACCAGTTCTAAAAGCTGTATTTGAGTTTATCACCCTTGTCACTAAGAAAAT
AAATGCAGGGTAAAATTTATATCCTTCTTGTTTTATGTTTCGGTATAAAACACTAATATC
AATTTCTGTGGTTATACTAAAAGTCGTTTGTTGGTTCAAATAATGATTAAATATCTCTTT
TCTCTTCCAATTGTCTAAATCAATTTTATTAAAGTTCATTTGATATGCCTCCTAAATTTT
TATCTAAAGTGAATTTAGGAGGCTTACTTGTCTGCTTTCTTCATTAGAATCAATCCTTTT
TTAAAAGTCAATATTACTGTAACATAAATATATATTTTAAAAATATCCCACTTTATCCAA
TTTTCGTTTGTTGAACTAATGGGTGCTTTAGTTGAAGAATAAAGACCACATTAAAAAATG
TGGTCTTTTGTGTTTTTTTAAAGGATTTGAGCGTAGCGAAAAATCCTTTTCTTTCTTATC
TTGATAATAAGGGTAACTATTGCCGTCGACTTTCGTCAGGGGGGCTTTTATTTATTCAAT
AATCCCTCCTCTCAATAAATCTATTGTTGTACTTAATTCAACTTCCATTTCTCTGTATCT
TTCAATACGCTCTTTTAAGTCCTTAATTTCTTTTTTTAATTCCTCATTTTCAGCAAATAA
CTCTTTTTCTTTGTTTGTCATTTTATTTCCCCCGTTTCAGCATCAAGAACCTTTGCATAA
CTTGCTCTATATCCACACTGATAATTGCCCTCAAACCATAATCTAAAGGCGCTAGAGTTT
GTTGAAACAATATCTTTTACATCATTCGTATTTAAAATTCCAAACTCCGCTCCCCTAAGG
CGAATAAAAGCCATTAAATCTTTTGTATTTACCAAATTATAGTCATCCACTATATCTAAA
AGTAAATTCTTCAATTCTCTTTTTTGGCTTTCATCAAGTGTTATATAGCGGTCAATATCA
AAATCATTAATGTTCAAAATATCTTTTTTGTCGTATATATGTTTATTCTTAGCAATAGCG
TCCTTTGATTCATGAGTCAAATATTCATATGAACCTTTGATATAATCAAGTATCTCAACA
TGAGCAACTGAACTATTCCCCAATTTTCGCTTAATCTTGTTCCTAACGCTTTCTATTGTT
ACAGGATTTCGTGCAATATATATAACGTGATAGTGTGGTTTTTTATAGTGCTTTCCATTT
CGTATAACATCACTACTATTCCATGTATCTTTATCTTTTTTTTCGTCCATATCGTGTAAA
GGACTGACAGCCATAGATACGCCCAAACTCTCTAATTTTTCTTTCCAATCATTAGGAATT
GAGTCAGGATATAATAAAAATCCAAAATTTCTAGCTTTAGTATTTTTAATAGCCATGATA
TATTACCTTATCAAAAACAAGTAGCGAAAACTCGTATCCTTCTAAAAACGCGAGCTTTCG
CTTATTTTTTTTGTTCTGATTCCTTTCTTGCATATTCTTCTATAGCTAACGCCGCAACCG
CAGATTTTGAAAAACCTTTTTGTTTCGCCATATCTGTTAATTTTTTATCTTGCTCTTTTG
TCAGAGAAATCATAACTCTTTTTTTCGATTCTGAAATCACCATTTAAAAAACTCCAATCA
AATAATTTTATAAAATTAGTGTATCACTTTGTAATCATAAAAACAACAATAAAGCTACTT
AAATATAGATTTATAAAAAACGTTGGCGAAAACGTTGGCGATTCGTTGGCGATTGAAAAA
CCCCTCAAACCCTTGAGCCAGTTGGGATAGAGCGTTTTTGGCACAAAAATTGGCACTCGG
CACTTAATGGGGGGTCGTAGTACGGAAGCAAAATTCGCTTCCTTTCCCCCCATTTTTTTC
CAAATTCCAAATTTTTTTCAAAAATTTTCCAGCGCTACCGCTCGGCAAAATTGCAAGCAA
TTTTTAAAATCAAACCCATGAGGGAATTTCATTCCCTCAAACTCCCTTGAGCCTCCTCCA
ACCGAAATAGAAGGACGCTGCGCTTATTATTTCATTCAGTCATCGGCTTTCATAATCTAA
CAGACAACATCTTCGCTGCAAAGCACGCTACGCTCAAGGGCTTTTACGCTACGATAACGC
CTGTTTTAACGATTATGCCGATAACTAAACGAAATAAACGCTAAAACGTCTCGGACATTC
TGCTCCCGCCCTTATGGGATTTATCTTCCTTATTCTCGCTTTGATTGTTCTATCGAAAGC
GAAATCAAACGAATAATTTATAAATAAAAAAACCACCTCTAAAAGGTGATTTTTATTTAT
AAATTACAATTTTTCAGCAATATCAGTAATTGCTTTATCAACTGCTGCTTTTTGGCTATC
AATCAAAGCAACACGTGCTGTAATTTGTTTAATTGCCATTTCAATTGAACGTTTCAAGCC
TTGGTTTTCTAATTTTGGTTCAAAGAAAGCTTGAGCTCTCTAGAACTAGTGGTAC
FolP
ATTTATGGCATCATGAATATTACGCCGGACTCCTTTTTCGACGGTGGCCAGTACAAGACT
AAAGATGACGTACTGAATCACGTGGATGCCATGTTACAAGCAGGAGCTGACGTAATCGAA
GTTAATGGTCAAACGACACGGCCGGGCTTCACGGAAGTCACGCCGCAAGTTGAATTAGAC
CGGACGTTGCCGTACATTCGCGCCATCAAGGCTCGTTTCCCGGAAGCAGTCTTAGCGGTC
GATACTTATAAGTATGATGTCATGCAAGCCGTCCTGGAAGAAGGTGTCAGCATTATTAAC
GATGTGAACGCCTTTACGGACGACCCTCGCAAGCTCAAATTGATGGCTGACAGTCGCGTC
GGCTTACTGACTATGCACAGTAGTCGTGACCAAGAATATACCGATCTGACGAGCAGTATG
CGTGGTTTCTTTGAACAAAATCTGGCAGCATTGACGAGTCATGGCATCGATATTGAACGA
ATTGCTCTAGATCAGGGAATCGGCTATTCACAAGTCGCACATGGGGAACAGGATTACGTG
ATGATGCGTAACATTGATGAATTTAATTATTTGCGACGCCCAATGATGGTGGCCATTTCA
CGGAAAGGTTATTTAGGTCTATTGCTTGGTCTGAAGAAGGAAGACCGGCTACCAATGACG
TTGGTCACTGAAGCAGCGATGATGCTTAAGGGTGGCCGGATCATCCGGGTCCACGATGTG
GCTGAAACCAAGCAATTGATTACGCTGTTAGGCCGAATTGAGAATGGCTATTGGTTATCG
AATAATGATTAG
Restriction sites: XbaI (486), ApoI (559), MfeI (733)
Xtp2
ATGACGACGACTTGGTTGATTGCATCCAACAACGCTGGTAAAAGCCGCGACTTGATCGCG
TGTTTGGCTTACTATGGCTTGACTGCTCGTCAGTACTTGACAGTGGCGCCGCGGCTCGAA
TTTCCCGTGGAAACGACGACGAGCTATGTCGATAACGCGGTTGCTAAAGCTCGTTTTGGG
GCGCAACAGCTAGGGGTTCCGGTTATCGCAGATGATAGTGGCTTAGAGATTTCCGCGTTA
CCAGACTTGTTAGGTGTGACCACGGCGCGCGACTTAGGGGTTGCAGTCAGTGGCTTTGAT
TGCAATCAGGAAATTTTAACGGCCCTACGCGATATCCCTGACAACGAGCGGCAAGCGTTG
ATGCGTGCTACGTTAGCGGCTGCCTGGCCAGATGGGCGGATCTTGGCCGTACAAGCTTCG
ATCACCGGCTACATTGCGTCCTATCAATTTGGACGCTATTCTGGGGGATTTGACCGAATT
TTCTGGCTACCGCGCTATGGTCGAACTTTGGCCGAATTACCAGCAACGTGGCGCATCCCG
CTGACACACCGGGGACGGGCCGCCTTAAAATTAATCACAAAACTTTAA
Restriction sites: none
FolC2
GTGGATACAGGGACGATTGAACAACGTTATCAGGACTTATTAGCCCAACTAAATCAAGCC
ATGTTAGCCAATAATCATCAGCGTGTTCCGTTATTACGGCGCATCATGGCACATCTTGGC
CACCCTGACCATTATTACCATGTGATCCATATCGCTGGGACTAATGGCAAAGGCTCTACG
GGAGCCATGTTAGCCAGTATCCTGCGGGCGCAAGGGTATCAAGTTGGCCGCTTCAGCAGC
CCCGCGATTAATGATGCGCGTGAACAACTGCAATGCAATGGGACGTGGATCAGTCCAGCG
GAATTTATCGATACGTATCGTGAGATTCTACCGGTTCTGCAAAATATGGGACTGACAGCT
AGCGATGTCTCCATCTTTGAATGGTTCTTTCTCATTAGTGTCGTTTGGTTTCGGAATCAA
AACGTGCAATGGGCGGTCATTGAAGCCGGTTTGGGCGGCTTGTATGATGCGACTAACGCC
TTAGCCAGTCCCCAACTCACGGTATTTACTAAGATTGCCCTCGATCATACCGCCATTCTT
GGGCCCACGATTACCGCGATTGCGCAAAATAAGTCAAAAATCATCAAGCCCCATACAACA
GCAGTGACATTGGCTGACCAACACCCAGAAGCCCTGGCTGTTTTGCAGACCGAAGCGCTC
AATCAAGGTGTTCGGCTAGTGACCGCTAAGCACGCGCAACTGACGGTGACTGGGCAAACA
CTCACGCAAACCGTCGTTGATGCGCACAGTCAGCTTTTTGATTGGACGCAATTAACGGTC
GGACTGAGTGGCACCTATCAGCTACAAAATCTGCGGTTGGTTTTAACTGTGGTTGCAGTA
CTACAACAGCAACAAGTTAACTTGACGAACAGTGCGGTTCGCCGAGGCTTGCAACAAGTT
TCCTTGCCAGGCCGGTTGACCGTATTACAGGAGCAACCTGTCATTATTGCCGATGGGGCG
CATAATCCGGATGGTATGGCGGCACTAGTGGCAAGTGTGAAGCAATTACTCGCTGGTCGT
AAATTGATTTGGATCGTCGGGGTCTTACAAGATAAGGCCTACCCTAAAATGCTGGCTGAA
CTATTACCGGTAGCTGACGTCCTGATTACGAATACGCCCGCAAATCCAGAACGAGCGTTA
TCCGCGACTAAGCTGGCCGACACGGCGAACGCATTATTAACGCCCACCTTTATGACCACG
GCTGCGATGCAACGACAGCCAGACTTATTGACTGCAACGACGATCACGGATGCGTTAACG
CAAGCTCAGCAGCAAGCAACCAGTAATAGTGCGATTATTGTAACGGGGTCATTTTATGTC
ATTCGTGAGCTCCAACAGGCTGGTTGGCCGGGGTTAAGCCAATGA
Restriction sites: ApoI (301), NheI (358), SpeI (984)
FolE
ATGATTGATGAGAAGAACCAAGCAAAGATTGAGCATGCAGTACGAGAAATTTTAAGTGCA
GTTGGTGAAGATCCTGACCGACCGGGATTAGTAGAAACACCAGCACGGGTTGCACGAATG
TATGCGGAAGTTTTCGCTACTAAGACTGCCGCACCATTTGATAATTATAAACTGTTCAAG
GTTGAGCATCCGACTGAAATGGTATTACTTAAGGATATTCCATTCTATTCGATGTGTGAG
CATCACCTTTTGCCGTTTTTTGGCACGGTTCAAGTTGCTTATGTGCCGCAGCATGAACAA
GTGATTGGCTTGAGTAAGATTCCTCGCTTGATTGACTATTGCAGTCAACAGCCGAACGTT
CAGGAGCGGTTGACAGTTTCCATTGCAACAGAATTACAACGAATTCTTGACCCGGCTGGG
ATCGCGGTCTCAATCACGGCGCGGCACATGTGCATGGAGATGCGGGGTGTTAGCAAACCG
GGTGTGCATACGGAAAGTAGCTATTACAGTGGTCAATTCAAGACGGATTTAGACTTGAAA
CGAGAATTCTTACAGCGAATCGCAAAGTAG
Restriction sites: none
FolK
ATGGCAAGTAGGGAAGAACGGGTTTATTTGAGTGTTGGTTCCAATATTCATCCGCGCGTC
CAAAATATTCAGCAAGCCCTTAGCCGATTACGAGCCGTCAATGGGGTAAACGTGATTGAC
GAATCTCATTGGTATGAGACTCAACCGTGGGGAAAGCGTGATCAGGCCAATTTTTACAAT
GTTTCGGTATCCTTAACGACTAATTTGACACCAGAAGAACTATTGGATGAATTACATACA
ATTGAGCAGGCGGGCCACCGCCAACGCTTGGTTCACTGGGGACCACGTACGATTGATTTG
GATATTATTTTTTGGGGCGACCGGCAAATCAACACAGCGACGCTGACGATTCCGCATGCG
CAGGCAGCTAAGCGCAACTTTGTGCTACTGCCAACTGCTGAAATCGCCAAAACTGATGTG
TTAGTTGGACCACAAGTGGCCCAATTGATTGCGGCTAATCAGGATCAGAGTTGGATTAAA
AAAGTAAGAAATGTGAGTGAGTTAGATGATTGA
Restriction sites: none
FolB
ATGGGCATGATTCGAATTAATAATTTACGCTTTCACACGTTTAACGGGGTACTTCCGGAA
GAACGGCGTAATGGTCAACAACTAGGGCTAGATATTGCCATTAAATATCCTATCGAAACC
AAGGTTCAACACGATGACGTTCACGAGACCATCAATTACGCGGCGGTCCGTAACGTGGTC
GATGAATTTGTAACGACCCATTCATACAAGTTGATTGAATCGCTAGCTAACCACTTATTG
CAGACGTTATTGACAAGTTTTCCCGCGGCGGATGCAATCAATATTAAAATTCGTAAATAT
AGCGTACCAATGCCTGGAATCTTTGATGATGTGGAAATTGAGGTGGAGGGGACGCCGAAT
GGCAAGTAG
Restriction sites: NheI (222)
Tyrosinase
DNA Sequence - http://partsregistry.org/wiki/index.php?title=Part:BBa_K193600
Contains gap at around 885 bp, causing a frame-shift
ATGGCGTGGCTGGTCGGCAAGCCGTCGCTCGAACGATCATGGAATGCGATACTAAGTTTT
CCGGAATCTGGATTCCAACTGGAATGCAGGAATACGATCGGTAGTAGCGTTTTTAGCTCG
CATTTTACGCTGCATTTTCGAGTGGCACGTCGCTTGCTTCATTTCTCGTGCCGCCGATTC
ACCGAAACTCAGAAAGAGCCAACGCAAGCTCTATGGTGGTGCGAATTGCCCACTGCTCCC
GCGCCCAGACGCCGCGGAACCGGCCTAAAAGCGGCATTAATCTTGGCGAAGGACAATTCA
AATCCAAGGGAGAGTAAGATGAGCATCACACGCAGACATGTCATCGTTCAGGGTGGCGTC
ATTGCAGCAGGCCTGCTCGCCAGCGGCCTACCGGGGACAAAAGCCTTCGCGCAGATACCG
TCAATCCCTTGGCGGCGCTCACTCCAGGGCTTGGCCTGGAACGACCCGATCATCGAGACC
TATCGCGACGCAGTGCGCCTTCTCAACGCCCTTCCCGCCAGCGACAAATTCAACTGGGTC
AACCTCTCGAAAATTCACGGCAGCGGTGACGTCGTCAAATACTGCCCGCATGGGAACTGG
TATTTCCTGCCGTGGCACAGGGCCTATACCGCTATGTACGAGCGCATCGTTCGGCACGTG
ACCAAGAACAACGATTTCGCTATGCCGTTCTGGGACTGGACCGACAATCCGTACCTGCCC
GAAGTGTTCACAATGCAAAAGACGCCCGACGGCAAGGACAATCCACTTTATGTTTCGTCG
CGCACCTGGCCAATCACGCAGCCGATGCCGGACAATATAGTTGGGCCACAGGTTCTCAAC
ACCATCCTAACGGCGAAGCCATACGAGGTCTTCGGCACCACCCGCCCGAGGGACAGAACT
CACTCGATCCTTCCTGGGTCACCACCAGCAGCGGCACGCAGGGGGCGCTGGAATACACAC
CGCACAATCAGGTGCACAACAATATCGGTGGCTGGATGCCGGAAATGTCGTCGCCCCGCG
ATCCGATCTTCTTCATGCATCATTGCAACATCGACCGCATCTGGGCGACCTGGAATTTGC
GCAACGCCAACAGCACGGATCGACTCTGGGCCGACATGCCGTTCACCGACAATTTCTACG
ATGTAGACGGCAACTTCTGGTCCCCGAAGGTCTCTGACCTTTATGTTCCAGAGGAACTCG
GATACAATTATGGTTTCCGGACCTACTTCAAGGTCGCGGCGGCGAGCGCCAAAACGCTGG
CCCTGAACGATAAACTCACGTCCGTGATCGCGGCGACGGCGACCGATGCTGCAATCGCCG
GCGTGACGACCACCTCCACGGACAACAGCAAGGCGGCAACGGAAAACGTGCCGCTTTCGC
TGCCGATCAAGATCCCGGCGGGCGCATTGCAGGAGATCGTCCGCCAACCGCCTCTGCCAT
CCGGCATGGATACAATGGATTTCGGCGCGGCACAGGAGCAGGCGGCCTCCGCTCCTCGTG
TGCTGGCATTCCTGCGCGATGTCGAGATCACCAGTGCCAGCACGACCAGCGTTCGGGTGT
TTCTCGGCAAGAACGACCTTAAGGCCGATACGCCCGTCACCGATCCCCATTACGTCGGTT
CTTTCGCCGTTCTCGGTCATGACGGCGACCATCATCGCAAACCATCCTTCGTCCTCGATC
TGACGGACGCGATCCAGCGGGTTTACGGCGGAAGGGGGCAGACGGATGGCGAGGCCATCG
ACCTCCAGCTCATTCCTGTCGGATCGGGAGCGGGCAAACCCGGCGCCGTAGAGCCCGCAA
AGCTAGAAATAGCCATAGTGTCCGCCTAA
Restriction sites: NsiI (1035), NgoMIV (1317)
Catalase
-
Superoxidase dismutase
(dimer without unspecific crosslinking)
C6A & C11A (crosslinking)
ATGGCGACGAAGGCCGTGGCCGTGCTGAAGGGCGACGGCCCAGTGCAGGGCATCATCAAT
TTCGAGCAGAAGGAAAGTAATGGACCAGTGAAGGTGTGGGGAAGCATTAAAGGACTGACT
GAAGGCCTGCATGGATTCCATGTTCATGAGTTTGGAGATAATACAGCAGGCTGTACCAGT
GCAGGTCCTCACTTTAATCCTCTATCCAGAAAACACGGTGGGCCAAAGGATGAAGAGAGG
CATGTTGGAGACTTGGGCAATGTGACTGCTGACAAAGATGGTGTGGCCGATGTGTCTATT
GAAGGTTCTGTGATCTCACTCTCAGGAGACCATGCCATCATTGGCCGCACACTGGTGGTC
CATGAAAAAGCAGATGACTTGGGCAAAGGTGGAAATGAAGAAAGTACAAAGACAGGAAAC
GCTGGAAGTCGTTTGGCTTGTGGTGTAATTGGGATCGCCCAATAA
Restriction sites: none
yCCS (yeast copper chaperone)
ATGACCACGAACGATACATACGAGGCTACTTATGCCATTCCCATGCACTGTGAAAATTGC
GTGAATGATATAAAGGCATGCCTAAAAAATGTCCCAGGAATCAATTCATTGAATTTCGAT
ATAGAGCAACAAATAATGAGCGTGGAAAGCTCAGTAGCCCCTTCAACCATCATCAACACC
TTGCGAAACTGTGGTAAAGACGCCATCATAAGAGGCGCCGGGAAGCCGAATTCTTCCGCG
GTCGCTATCTTAGAGACCTTTCAAAAATATACTATAGACCAGAAAAAGGACACCGCCGTG
AGGGGCCTAGCAAGAATCGTTCAGGTTGGAGAGAATAAGACTCTTTTTGACATCACTGTA
AATGGCGTCCCAGAGGCGGGAAATTACCACGCCAGCATTCACGAGAAAGGCGACGTCTCC
AAGGGCGTGGAATCTACTGGTAAAGTTTGGCACAAGTTCGACGAACCCATTGAATGCTTC
AACGAGAGTGATTTGGGCAAGAACTTGTACAGTGGGAAGACCTTTCTGAGCGCTCCCTTG
CCAACATGGCAACTTATCGGCCGCAGCTTTGTCATTTCCAAGTCTCTGAACCACCCAGAA
AACGAGCCTTCCTCCGTTAAGGACTACTCATTTCTGGGAGTCATTGCGAGAAGCGCTGGT
GTGTGGGAAAATAATAAACAAGTGTGTGCGTGTACCGGAAAGACTGTTTGGGAAGAGAGA
AAAGATGCCTTGGCCAACAACATCAAATAG
Restriction sites: EcoRI (228)
bFGF
ATGGCAGCCGGGAGCATCACCACGCTGCCCGCCTTGCCCGAGGATGGCGGCAGCGGCGCC
TTCCCGCCCGGCCACTTCAAGGACCCCAAGCGGCTGTACTGCAAAAACGGGGGCTTCTTC
CTGCGCATCCACCCCGACGGCCGAGTTGACGGGGTCCGGGAGAAGAGCGACCCTCACATC
AAGCTACAACTTCAAGCAGAAGAGAGAGGAGTTGTGTCTATCAAAGGAGTGTGTGCTAAC
CGTTACCTGGCTATGAAGGAAGATGGAAGATTACTGGCTTCTAAATGTGTTACGGATGAG
TGTTTCTTTTTTGAACGATTGGAATCTAATAACTACAATACTTACCGGTCAAGGAAATAC
ACCAGTTGGTATGTGGCACTGAAACGAACTGGGCAGTATAAACTTGGATCCAAAACAGGA
CCTGGGCAGAAAGCTATACTTTTTCTTCCAATGTCTGCTAAGAGCTGA
Restriction sites: AgeI (344)
MGF
-
MITF
GenBank: Z29678.1
>gi|468496:121-1380 H.sapiens mitF mRNA
ATGCTGGAAATGCTAGAATATAATCACTATCAGGTGCAGACCCACCTCGAAAACCCCACC
AAGTACCACATACAGCAAGCCCAACGGCAGCAGGTAAAGCAGTACCTTTCTACCACTTTA
GCAAATAAACATGCCAACCAAGTCCTGAGCTTGCCATGTCCAAACCAGCCTGGCGATCAT
GTCATGCCACCGGTGCCGGGGAGCAGCGCACCCAACAGCCCCATGGCTATGCTTACGCTT
AACTCCAACTGTGAAAAAGAGGGATTTTATAAGTTTGAAGAGCAAAACAGGGCAGAGAGC
GAGTGCCCAGGCATGAACACACATTCACGAGCGTCCTGTATGCAGATGGATGATGTAATC
GATGACATCATTAGCCTAGAATCAAGTTATAATGAGGAAATCTTGGGCTTGATGGATCCT
GCTTTGCAAATGGCAAATACGTTGCCTGTCTCGGGAAACTTGATTGATCTTTATGGAAAC
CAAGGTCTGCCCCCACCAGGCCTCACCATCAGCAACTCCTGTCCAGCCAACCTTCCCAAC
ATAAAAAGGGAGCTCACAGCGTGTATTTTTCCCACAGAGTCTGAAGCAAGAGCACTGGCC
AAAGAGAGGCAGAAAAAGGACAATCACAACCTGATTGAACGAAGAAGAAGATTTAACATA
AATGACCGCATTAAAGAACTAGGTACTTTGATTCCCAAGTCAAATGATCCAGACATGCGC
TGGAACAAGGGAACCATCTTAAAAGCATCCGTGGACTATATCCGAAAGTTGCAACGAGAA
CAGCAACGCGCAAAAGAACTTGAAAACCGACAGAAGAAACTGGAGCACGCCAACCGGCAT
TTGTTGCTCAGAATACAGGAACTTGAAATGCAGGCTCGAGCTCATGGACTTTCCCTTATT
CCATCCACGGGTCTCTGCTCTCCAGATTTGGTGAATCGGATCATCAAGCAAGAACCCGTT
CTTGAGAACTGCAGCCAAGACCTCCTTCAGCATCATGCAGACCTAACCTGTACAACAACT
CTCGATCTCACGGATGGCACCATCACCTTCAACAACAACCTCGGAACTGGGACTGAGGCC
AACCAAGCCTATAGTGTCCCCACAAAAATGGGATCCAAACTGGAAGACATCCTGATGGAC
GACACCCTTTCTCCCGTCGGTGTCACTGATCCACTCCTTTCCTCAGTGTCCCCCGGAGCT
TCCAAAACAAGCAGCCGGAGGAGCAGTATGAGCATGGAAGAGACGGAGCACACTTGTTAG
Restriction sites: AgeI (189), PstI (969)
Rare codons
When overexpressing genes from other organisms. Initially we won't be very worried about this, however there might be lower expression of the protein from the bacteria we will use. If this becomes a bigger problem we might use bacteria strains that already contains tRNA for different rare codons.
Neglect, synthesize the gene, overexpress rare tRNA or use a strain that already overexpress the tRNA
Search on e.g. http://nihserver.mbi.ucla.edu/RACC/
- IgG protease
http://korta.nu/053b 5 rare codons for arginine, 9 for leucine, 4 for isoleucine, 1 for proline
- Melanin
http://korta.nu/053b 36 rare codons for arginine, 5 for leucine, 7 for isoleucine, 10 for proline
Nina: Even though melanin looks like it has many rare codons, we should remember that team Tokyo Tech 09 had melanin expressed in e.coli, which then leaked out melanin. Therefore it seems that we might not be too bothered about this many rare codons.
Johan Yes, Tokyo Tech 2009 used the normal K-12 JM109 strain. If a gene still has too many rare codons, it seems like we can use the rosetta strains on http://openwetware.org/wiki/E._coli_genotypes
Primer design information
To create our constructs when we have the genes and their sequences
Several strategies for creating and joining two independent DNAs without the use of restriction endonucleases have been developed. One such strategy uses overlap extension employing two PCR steps (7–10). Two intermediate PCR fragments with overlapping end sequences are first amplified, then re-annealed across the overlapping sequences, extended and subsequently amplified with a flanking primer set (7). Though the idea is attractive, the success of overlap PCR depends on efficient cross- annealing of intermediate PCR products and effective removal of unused internal primers to direct the second amplification of the full-length product. - USER TM friendly DNA engineering and cloning method by uracil excision
[< rev]
[leading strand] 5' --------------------------------------------- 3'
[lagging strand] 3' --------------------------------------------- 5'
[forw >]
Note! We will use iGEM team Freiburg 2007 fusion parts (BBF RFC 25) as our assembly standard 25 and thus their prefix and suffix, which differ from the standard prefix and suffix.
https://2007.igem.org/Freiburg07/report_fusion_parts
http://openwetware.org/wiki/The_BioBricks_Foundation:Standards/Technical/Formats#The_Berkeley_.28BBb.29_Format scroll down to BBF RFC 25.
The "mixed site/scar" of the standard prefix and suffix would result in an in-frame shift and also a stop codon when fusing two proteins together, such as in constructing a fusion protein. When making fusion proteins with the Freiburg 2007 prefix and suffix the "mixed site/scar" will be in frame and also not containing any stop codons. This is the reason we will use the team Freiburg 2007 fusion parts.
[forw >]
5' [extra base pairs] [biobrick prefix] [beginning of insert] 3'
~3-6 nt 20/22 nt ~18-22 nt
[< rev]
5' [extra base pairs] [biobrick suffix] ([extra TTA]) [end of insert] 3'
~3-6 nt 21 nt 3 nt ~18-22 nt
- [extra base pairs] : Needed for the restriction enzymes.
Should start with G/C for end stability (for the later cycles of the PCR).
Andreas used GAA.
Extra information on http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/cleavage_linearized_vector.asp
- [biobrick prefix] : GAATTCGCGGCCGCTTCTAGATGGCCGGC (29 nt)
(http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix).
- [biobrick suffix] : ACCGGTTAATACTAGTAGCGGCCGCTGCAG (30 nt)
(http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix) Make this reverse and complementary in the primer design.
- Constructs
To send to iGEM as biobrick-parts:
- prefix-CPP-suffix
- prefix-Z-suffix
- prefix-IgG protease-suffix
- prefix-Tyrosinase-suffix
- prefix-SOD-suffix
- prefix-yCCS-suffix (coppar chaperone for SOD)
- prefix-MITF-suffix
- prefix-bFGF-suffix
- prefix-(vitamin B cluster)-suffix
- prefix-His-suffix
To make additionaly:
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-Z-suffix)-(prefix-IgG-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-Tyrosinase-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-SOD-suffix)-(prefix-yCCS-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-Tyrosinase-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-MITG-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)-(prefix-bFGF-suffix)-(prefix-His-suffix)
- (prefix-Signal-suffix)-(prefix-CPP-suffix)- ( (prefix-vitamin-B-cluster-suffix) )-(prefix-His-suffix)
Assembly standard 25
http://mbcf.dfci.harvard.edu/docs/oligocalc.html
* Vitamin B cluster
* Mitf
- IgG protease:
F: 5'- GATG GCCGGC GATAGTTTTTCTGCTAATCAAGAGATTA
Early cycles: 28 nt, 53 °C, 29 %
Later cycles: 62 nt, 74 °C, 48 %
R: 5'- ATTA ACCGGT ATTGGTCTGATTCCAACTATCTTGC
Early cycles: 25 nt, 54 °C, 40 %
Later cycles: 60 nt, 75 °C, 50 %
- Protein A, Z domain:
F: 5'- GATG GCCGGC GTAGACAACAAATTCAACAAAGAACAAC
Early cycles: 28 nt, 54 °C, 32 %
Later cycles: 62 nt, 75 °C, 50 %
R: 5'- ATTA ACCGGT TTTCGGCGCCTGAGCATCAT
Early cycles: 20 nt, 54 °C, 55 %
Later cycles: 56 nt, 75 °C, 52 %
- SOD:
Second PCR:
F: 5'- GATG GCCGGC GCGACGAAGGCCGTG
Early cycles: 15 nt, 55,7 °C, 73 %
Later cycles: 50 nt, 78 °C, 64 %
R: 5'- ATTA ACCGGT TTGGGCGATCCCAATTACACC
Early cycles: 21 nt, 54 °C, 52 %
Later cycles: 57 nt, 74 °C, 51 %
- Tyrosinase:
F: 5'- GATG GCCGGC GCGTGGCTGGTCGGCAAG
Early cycles: 18 nt, 57 °C, 72 %
Later cycles: 52 nt, 79 °C, 63 %
R: 5'- ATTA ACCGGT GGCGGACACTATGGCTATTTCTAG
Early cycles: 24 nt, 57 °C, 50 %
Later cycles: 59 nt, 75 °C, 51 %
- yCCS:
F: 5'- GATG GCCGGC ACCACGAACGATACATACGAGGC
Early cycles: 23 nt, 57 °C, 52 %
Later cycles: 57 nt, 77 °C, 56 %
R: 5'- ATTA ACCGGT TTTGATGTTGTTGGCCAAGGCATC
Early cycles: 24 nt, 56 °C, 46 %
Later cycles: 59 nt, 74 °C, 49 %
- bFGF:
F: 5'- GATG GCCGGC GCAGCCGGGAGCATCACC
Early cycles: 18 nt, 60,5 °C, 72 %
Later cycles: 51 nt, 78 °C, 63 %
R: 5'- ATTA ACCGGT GCTCTTAGCAGACATTGGAAGAAAAAGT
Early cycles: 28 nt, 58,1 °C, 39 %
Later cycles: 58 nt, 73 °C, 48 %
To be synthesized:
- Cell penetrating peptide (CPP)
NgoMI-( ATG CPP )-AgeI
NgoMI cutting: http://www.neb.com/nebecomm/productfiles/423/images/NgoM-IV-cutsite_1_v1_000017.gif
AgeI cutting: http://www.neb.com/nebecomm/productfiles/28/images/Age-I-cutsite_1_v1_000017.gif
- LMWP
Final sequence: GATG GCCGGC ATGGTTTCTCGTCGTCGTCGTCGTCGTGGTGGTCGTCGTCGTCGT ACCGGT TAAT
Sequence of the four oligos:
GATG GCCGGC ATGGTTTCTCGTCGTCG TCGTCGTCGTGGTGGTCGTCGTCGTCGT ACCGGT TAAT
CTAC CGGCCG TACCAAAGAGCAGCAGCAGCAGC AGCACCACCAGCAGCAGCAGCA TGGCCA ATTA
- Oligo 1: CCGGC ATGGTTTCTCGTCGTCG
- Oligo 2: TCGTCGTCGTGGTGGTCGTCGTCGTCGT A
- Oligo 3: G TACCAAAGAGCAGCAGCAGCAGC
- Oligo 4: AGCACCACCAGCAGCAGCAGCA TGGCC
- Dowdys
Final sequence: GATG GCCGGC ATGTACGGTCGTAAAAAACGTCGTCAGCGTCGTCGT ACCGGT TAAT
Sequence of the four oligos:
GATG GCCGGC ATGTACGGTCGTAAAA AACGTCGTCAGCGTCGTCGT ACCGGT TAAT
CTAC CGGCCG TACATGCCAGCATTTTTTGCAG CAGTCGCAGCAGCA TGGCCA ATTA
- Oligo 1: CCGGC ATGTACGGTCGTAAAA
- Oligo 2: AACGTCGTCAGCGTCGTCGT A
- Oligo 3: G TACATGCCAGCATTTTTTGCAG
- Oligo 4: CAGTCGCAGCAGCA TGGCC
- Transportan 10:
Final sequence: GATG GCCGGC ATGGCGGGTTACCTGCTGGGTAAAATCAACCTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTG ACCGGT TAAT
Sequence of the four oligos:
GATG G CCGGC ATGGCGGGTTACCTGCTGGGTAAAATCAAC CTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTG ACCGGT TAAT
CTAC CGGCC G TACCGCCCAATGGACGACCCATTTTAGTTGGACTTTCGC GACCGCCGCGACCGCTTTTTTTAGGAC TGGCC A ATTA
- Oligo 1: CCGGC ATGGCGGGTTACCTGCTGGGTAAAATCAAC
- Oligo 2: CTGAAAGCGCTGGCGGCGCTGGCGAAAAAAATCCTG A
- Oligo 3: G TACCGCCCAATGGACGACCCATTTTAGTTGGACTTTCGC
- Oligo 4: GACCGCCGCGACCGCTTTTTTTAGGAC TGGCC
Primer test
This is for testing our designed primers. Primer-tests such as hairpin, self-dimer, etc will be done and also BLASTing the primers against the template to avoid unspecific bindning.
The test will be done at: http://eu.idtdna.com/analyzer/applications/oligoanalyzer/Default.aspx and also in the program AmplifX, which makes a prediction if one obtains a PCR product based on chosen primers.
- AmplifX:
- Cell penetrating peptide (CPP) -
- Protein A -
- IgG protease -
Amplified fragment size : 930
GC% : 37,0%
Suggested annealing temperature : 53,1
Forward primer : Pr0001
Reverse primer : Pr0002
Primer : Pr0001 : 5' GATAGTTTTTCTGCTAATCAAGAGATTA 3'
||||||||||||||||||||||||||||
Target : 4 : 5' GATAGTTTTTCTGCTAATCAAGAGATTA 3'
score : 184
TM : 52,0
Primer : Pr0002 : 3' GCAAGATAGTTGGAATCAGACCAAT 5' (complement)
|||||||||||||||||||||||||
Target : 909 : 5' GCAAGATAGTTGGAATCAGACCAAT 3'
score : 181
TM : 55,4
- Melanin -
- MITF (Microphthalmia-associated transcription factor) -
- Catalase -
- Superoxide dismutase -
- Mast cell growth factor (MGF)-
- Basic fibroblast growth factor (bFGF) -
- Vitamin D
- Vitamin B12 (cyanoocobalamin) -
- Vitamin B9 (folic acid) -
Vectors the genes come in (to better find e.g. unspecific binding to template)
- bFGF: pGEX-bFGF
- Vitamin B9 cluster: pNZ7026
- Protein A: ?
- MITF: pRc/CMV-MITF-M / pGEX-MITF
- IgG protease: pET28c
- yCSS/SOD: pACA-SOD1
- Tyrosinase: ?
 "
"