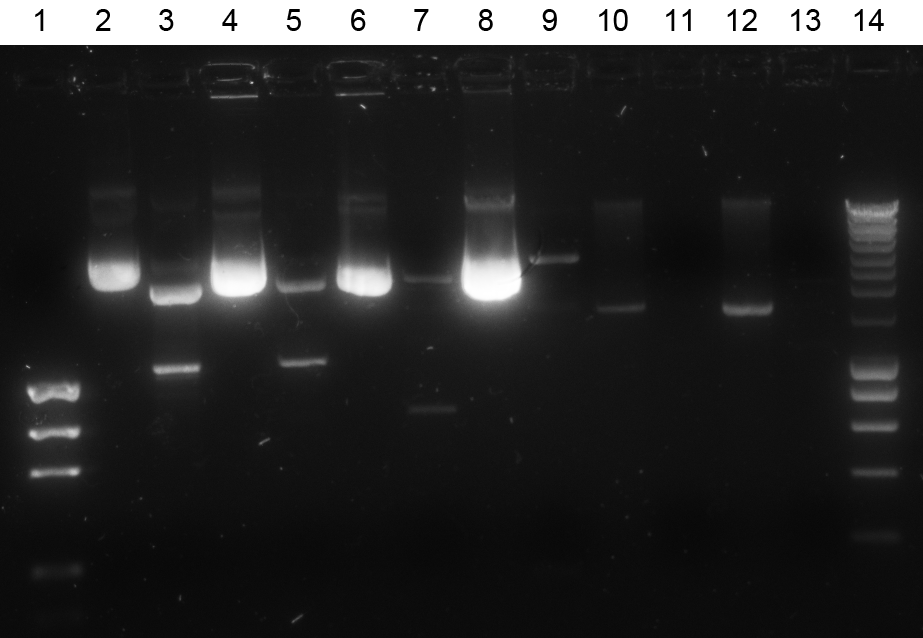Team:TU Delft/13 July 2010 content
From 2010.igem.org
(→Lab work) |
|||
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
=Lab work= | =Lab work= | ||
| - | + | ==Ordered DNA== | |
We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digested]] the plasmids: | We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we [[Team:TU_Delft/protocols/restriction_enzyme_digestion|digested]] the plasmids: | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''Sample''' |
| - | |''' | + | |'''Enzyme 1''' |
| + | |'''Enzyme 2''' | ||
| + | |'''Buffer''' | ||
| + | |'''BSA''' | ||
|'''Needed fragment''' | |'''Needed fragment''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |1 μg alkB2 | + | |1 μg alkB2 |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–alkB2–P’ | ||
|- | |- | ||
|2 | |2 | ||
| - | |1 μg rubA3 | + | |1 μg rubA3 |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–rubA3–P’ | ||
|- | |- | ||
|3 | |3 | ||
| - | |1 μg ladA | + | |1 μg ladA |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–ladA–P’ | ||
|- | |- | ||
|4 | |4 | ||
| - | |1 μg ADH | + | |1 μg ADH |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–ADH–P’ | ||
|- | |- | ||
|5 | |5 | ||
| - | |1 μg AlnA | + | |1 μg AlnA |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–AlnA–P’ | ||
|- | |- | ||
|6 | |6 | ||
| - | |1 μg OprG | + | |1 μg OprG |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–OprG–P’ | ||
|- | |- | ||
|7 | |7 | ||
| - | |1 μg AlkS | + | |1 μg AlkS |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–AlkS–P’ | ||
|- | |- | ||
|8 | |8 | ||
| - | |1 μg PalkB | + | |1 μg PalkB |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–PalkB–P’ | ||
|- | |- | ||
|9 | |9 | ||
| - | |1 μg PalkS12 | + | |1 μg PalkS12 |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–PalkS12-P’ | ||
|- | |- | ||
|10 | |10 | ||
| - | |1 μg PhPFDα | + | |1 μg PhPFDα |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–PhPFDα–P’ | ||
|- | |- | ||
|11 | |11 | ||
| - | |1 μg PhPFDβ | + | |1 μg PhPFDβ |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–PhPFDβ–P’ | ||
|- | |- | ||
|12 | |12 | ||
| - | |3 μg pSB1C3 | + | |3 μg pSB1C3 |
| - | | | + | |EcoRI |
| - | | | + | |PstI |
| + | |3 (BioLabs) | ||
| + | |✗ | ||
| + | |‘E–linear pSB1C3–P’ | ||
|} | |} | ||
| - | + | ==Solvent Tolerance and Hydrocarbon Sensing== | |
Some BioBricks are in production. [[Team:TU_Delft/protocols/restriction_enzyme_digestion|Digestion reaction]] | Some BioBricks are in production. [[Team:TU_Delft/protocols/restriction_enzyme_digestion|Digestion reaction]] | ||
{| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
| - | |''' | + | |'''Sample''' |
| - | |''' | + | |'''Enzyme 1''' |
| + | |'''Enzyme 2''' | ||
| + | |'''Buffer''' | ||
| + | |'''BSA''' | ||
|'''Needed fragment''' | |'''Needed fragment''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |1 μg PhPFDα | + | |1 μg PhPFDα |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–PhPFDα–S’ | ||
|- | |- | ||
|2 | |2 | ||
| - | |1 μg PhPFDβ | + | |1 μg PhPFDβ |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–PhPFDβ–S’ | ||
|- | |- | ||
|3 | |3 | ||
| - | |1 μg AlkS | + | |1 μg AlkS |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–AlkS–S’ | ||
|- | |- | ||
|4 | |4 | ||
| - | |1 μg PalkB | + | |1 μg PalkB |
| - | | | + | |EcoRI |
| - | | | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E–PalkB–S’ | ||
|- | |- | ||
|5 | |5 | ||
| - | |1 μg B0032 | + | |1 μg B0032 |
| - | | | + | |XbaI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–B0032–P’ | ||
|- | |- | ||
|6 | |6 | ||
| - | |1 μg B0015 | + | |1 μg B0015 |
| - | | | + | |XbaI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–B0015–P’ | ||
|- | |- | ||
|7 | |7 | ||
| - | |1 μg E0422 | + | |1 μg E0422 |
| - | | | + | |XbaI |
| - | | | + | |PstI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘X–E0422–P’ | ||
|- | |- | ||
|8 | |8 | ||
| - | |3 μg pSB1T3 | + | |3 μg pSB1T3 |
| - | | | + | |EcoRI |
| - | |‘E - | + | |SpeI |
| + | |2 (BioLabs) | ||
| + | |✓ | ||
| + | |‘E-linear pSB1T3-P’ | ||
|} | |} | ||
| - | + | ==Emulsifier== | |
Today the digestion products from [[Team:TU_Delft/12_July_2010|yesterday]] were run on gel to see whether the plasmids were cut in the right way. | Today the digestion products from [[Team:TU_Delft/12_July_2010|yesterday]] were run on gel to see whether the plasmids were cut in the right way. | ||
| - | [[Image:TU_Delft_2010-07-13_P_Digestion.png|thumb|left|600px|1% | + | [[Image:TU_Delft_2010-07-13_P_Digestion.png|thumb|left|600px|1 % agarose of digestion check. Gel runned 1 hour at 100 V. Of all samples 10 μL + 2 μL loadingbuffer was loaded and 5 μL was loaded of marker]] |
| - | + | Lane description: | |
| - | + | ||
| - | Lane description | + | |
| - | : | + | |
{|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
|'''#''' | |'''#''' | ||
|'''Description''' | |'''Description''' | ||
|'''Expected Length (bp)''' | |'''Expected Length (bp)''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
|- | |- | ||
|1 | |1 | ||
| - | |Biorad EZ marker | + | |Biorad EZ marker |
| + | |n/a | ||
| + | |n/a | ||
| | | | ||
|- | |- | ||
|2 | |2 | ||
| - | |Undigested pSB1T3 | + | |Undigested pSB1T3 |
| + | |3507 | ||
| + | |<font color=limegreen>✓</font> | ||
| | | | ||
|- | |- | ||
|3 | |3 | ||
| - | | | + | |pSB1T3 + EcoRI + PstI |
| - | | | + | |1085, 2422 |
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
|- | |- | ||
|4 | |4 | ||
| - | |Undigested AlnA | + | |Undigested AlnA |
| + | |3469 | ||
| + | |<font color=limegreen>✓</font> | ||
| | | | ||
|- | |- | ||
|5 | |5 | ||
| - | | | + | |AlnA + EcoRI + SpeI |
| - | | | + | |1071, 2398 |
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
|- | |- | ||
|6 | |6 | ||
| - | |Undigested OprG | + | |Undigested OprG |
| + | |3122 | ||
| + | |<font color=limegreen>✓</font> | ||
| | | | ||
|- | |- | ||
|7 | |7 | ||
| - | | | + | |OprG + EcoRI + SpeI |
| - | | | + | |708, 2414 |
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
|- | |- | ||
|8 | |8 | ||
| - | |Undigested B0015 | + | |Undigested B0015 |
| + | |3318 | ||
| + | |<font color=limegreen>✓</font> | ||
| | | | ||
|- | |- | ||
|9 | |9 | ||
| - | | | + | |B0015 + XbaI + PstI |
| - | | | + | |155, 3163 |
| + | |<font color=limegreen>✓</font> | ||
| + | |fragment probably run of the gel | ||
|- | |- | ||
|10 | |10 | ||
| - | |Undigested R0011 | + | |Undigested R0011 |
| - | | | + | |2134 |
| + | |<font color=limegreen>✓</font> | ||
| + | |Sample not fully loaded on gel | ||
|- | |- | ||
|11 | |11 | ||
| - | | | + | |R0011 = EcoRI + SpeI |
| - | | | + | |78, 2056 |
| + | |? | ||
| + | |fragment probably run of the gel | ||
|- | |- | ||
|12 | |12 | ||
| - | |Undigested B0032 | + | |Undigested B0032 |
| + | |2092 | ||
| + | |<font color=limegreen>✓</font> | ||
| | | | ||
|- | |- | ||
|13 | |13 | ||
| - | | | + | |B0032 + XbaI + PstI |
| - | | | + | |39, 2053 |
| + | |? | ||
| + | |fragment probably run of the gel | ||
|- | |- | ||
|14 | |14 | ||
| - | |SmartLadder | + | |SmartLadder |
| + | |n/a | ||
| + | |n/a | ||
| | | | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
The ligation products that were incubated over night were transformed to Top10 competent cells according to the [[Team:TU_Delft/protocols/transformation|protocol]]. | The ligation products that were incubated over night were transformed to Top10 competent cells according to the [[Team:TU_Delft/protocols/transformation|protocol]]. | ||
| Line 199: | Line 288: | ||
==Characterization of Anderson RBS sequences== | ==Characterization of Anderson RBS sequences== | ||
| - | The first attempt at measuring fluorescence was made in adherence with the [http://partsregistry.org/Part:BBa_K176011 | + | The first attempt at measuring fluorescence was made in adherence with the [http://partsregistry.org/Part:BBa_K176011 protocol] proposed by the USTC team of 2009: |
| + | |||
| + | 1. The overnight cultures of ''E.coli'' Top10 carrying K398500, K398501, K398502, K398503, K398504 and I13401 (LB, 37 ℃, 160 rpm) were measured for OD600 (see table below) and diluted 1:100. | ||
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''OD600''' | ||
| + | |- | ||
| + | |K398500 | ||
| + | |1.443 | ||
| + | |- | ||
| + | |K398501 | ||
| + | |1.410 | ||
| + | |- | ||
| + | |K398502 | ||
| + | |1.448 | ||
| + | |- | ||
| + | |K398503 | ||
| + | |1.414 | ||
| + | |- | ||
| + | |K398504 | ||
| + | |1.552 | ||
| + | |- | ||
| + | |I13401 | ||
| + | |1.451 | ||
| + | |} | ||
| + | |||
| + | 2. 100 μL and 300 μL aliquots of the diluted cultures were pipetted into 96-well plates in three-fold. | ||
| - | + | 3. The plate was incubated at 37 ℃ while shaking for 3 hours. | |
| - | + | 4. The fluorescence and OD600 were measured for 16.30 hours with intervals 10 minutes by means of the Biotek Synergy plate reader with the excitation filter set at 485nm and the emission filter at 520nm. LB medium was used as blank reference. | |
| - | + | Note: surprisingly no over night growth of the cultures was observed in M9 minimal medium containing glucose. This a larger volume of inoculate (in LB) was used for a second stab at over night growth in M9. | |
| - | Plasmid purification of overnight cultures carrying K398500, K398501, K398502, K398503, K398504 in pSB1A3 was performed using the | + | Plasmid purification of overnight cultures carrying K398500, K398501, K398502, K398503, K398504 in pSB1A3 was performed using the [[Team:TU_Delft/protocols/midi-prep_plasmid_isolation|Qiagen Midi-prep plasmid isolation kit]] yielding sufficient plasmid DNA. These BioBricks can in future be placed under control of a low to medium copy number plasmid for comparison with high copy-number plasmid expression. |
Latest revision as of 19:57, 5 August 2010
Contents |
Lab work
Ordered DNA
We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we digested the plasmids:
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg alkB2 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–alkB2–P’ |
| 2 | 1 μg rubA3 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–rubA3–P’ |
| 3 | 1 μg ladA | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–ladA–P’ |
| 4 | 1 μg ADH | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–ADH–P’ |
| 5 | 1 μg AlnA | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–AlnA–P’ |
| 6 | 1 μg OprG | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–OprG–P’ |
| 7 | 1 μg AlkS | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–AlkS–P’ |
| 8 | 1 μg PalkB | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PalkB–P’ |
| 9 | 1 μg PalkS12 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PalkS12-P’ |
| 10 | 1 μg PhPFDα | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PhPFDα–P’ |
| 11 | 1 μg PhPFDβ | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–PhPFDβ–P’ |
| 12 | 3 μg pSB1C3 | EcoRI | PstI | 3 (BioLabs) | ✗ | ‘E–linear pSB1C3–P’ |
Solvent Tolerance and Hydrocarbon Sensing
Some BioBricks are in production. Digestion reaction
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA | Needed fragment |
| 1 | 1 μg PhPFDα | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PhPFDα–S’ |
| 2 | 1 μg PhPFDβ | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PhPFDβ–S’ |
| 3 | 1 μg AlkS | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–AlkS–S’ |
| 4 | 1 μg PalkB | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E–PalkB–S’ |
| 5 | 1 μg B0032 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0032–P’ |
| 6 | 1 μg B0015 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–B0015–P’ |
| 7 | 1 μg E0422 | XbaI | PstI | 2 (BioLabs) | ✓ | ‘X–E0422–P’ |
| 8 | 3 μg pSB1T3 | EcoRI | SpeI | 2 (BioLabs) | ✓ | ‘E-linear pSB1T3-P’ |
Emulsifier
Today the digestion products from yesterday were run on gel to see whether the plasmids were cut in the right way.
Lane description:
| # | Description | Expected Length (bp) | Status | Remarks |
| 1 | Biorad EZ marker | n/a | n/a | |
| 2 | Undigested pSB1T3 | 3507 | ✓ | |
| 3 | pSB1T3 + EcoRI + PstI | 1085, 2422 | ✓ | |
| 4 | Undigested AlnA | 3469 | ✓ | |
| 5 | AlnA + EcoRI + SpeI | 1071, 2398 | ✓ | |
| 6 | Undigested OprG | 3122 | ✓ | |
| 7 | OprG + EcoRI + SpeI | 708, 2414 | ✓ | |
| 8 | Undigested B0015 | 3318 | ✓ | |
| 9 | B0015 + XbaI + PstI | 155, 3163 | ✓ | fragment probably run of the gel |
| 10 | Undigested R0011 | 2134 | ✓ | Sample not fully loaded on gel |
| 11 | R0011 = EcoRI + SpeI | 78, 2056 | ? | fragment probably run of the gel |
| 12 | Undigested B0032 | 2092 | ✓ | |
| 13 | B0032 + XbaI + PstI | 39, 2053 | ? | fragment probably run of the gel |
| 14 | SmartLadder | n/a | n/a |
The ligation products that were incubated over night were transformed to Top10 competent cells according to the protocol.
Characterization of Anderson RBS sequences
The first attempt at measuring fluorescence was made in adherence with the [http://partsregistry.org/Part:BBa_K176011 protocol] proposed by the USTC team of 2009:
1. The overnight cultures of E.coli Top10 carrying K398500, K398501, K398502, K398503, K398504 and I13401 (LB, 37 ℃, 160 rpm) were measured for OD600 (see table below) and diluted 1:100.
| # | OD600 |
| K398500 | 1.443 |
| K398501 | 1.410 |
| K398502 | 1.448 |
| K398503 | 1.414 |
| K398504 | 1.552 |
| I13401 | 1.451 |
2. 100 μL and 300 μL aliquots of the diluted cultures were pipetted into 96-well plates in three-fold.
3. The plate was incubated at 37 ℃ while shaking for 3 hours.
4. The fluorescence and OD600 were measured for 16.30 hours with intervals 10 minutes by means of the Biotek Synergy plate reader with the excitation filter set at 485nm and the emission filter at 520nm. LB medium was used as blank reference.
Note: surprisingly no over night growth of the cultures was observed in M9 minimal medium containing glucose. This a larger volume of inoculate (in LB) was used for a second stab at over night growth in M9.
Plasmid purification of overnight cultures carrying K398500, K398501, K398502, K398503, K398504 in pSB1A3 was performed using the Qiagen Midi-prep plasmid isolation kit yielding sufficient plasmid DNA. These BioBricks can in future be placed under control of a low to medium copy number plasmid for comparison with high copy-number plasmid expression.
 "
"