Team:ETHZ Basel
From 2010.igem.org
| Line 4: | Line 4: | ||
[[Image:Setup.jpg|thumb|400px|'''Figure 1.''' Setup to control ''E. coli'' movements. An automatized microscope images the E. lemming. A connected computer system detects and tracks the cells. The direction of movement of the E. lemming is compared to the desired direction defined by the user, e.g. with a joystick. If the direction of movement deviates too much from the desired direction, the digital controller induces tumbling by sending a red light pulse. Otherwise, tumbling is repressed by sending a far-red light pulse.]] | [[Image:Setup.jpg|thumb|400px|'''Figure 1.''' Setup to control ''E. coli'' movements. An automatized microscope images the E. lemming. A connected computer system detects and tracks the cells. The direction of movement of the E. lemming is compared to the desired direction defined by the user, e.g. with a joystick. If the direction of movement deviates too much from the desired direction, the digital controller induces tumbling by sending a red light pulse. Otherwise, tumbling is repressed by sending a far-red light pulse.]] | ||
| - | The | + | The core idea of our project is to control the movement (chemotaxis) of ''E. coli''- |
by means of light! In fact, we will alter the chemotactic pathway | by means of light! In fact, we will alter the chemotactic pathway | ||
either by substituting the receptor with a light-sensitive one or by | either by substituting the receptor with a light-sensitive one or by | ||
| Line 16: | Line 16: | ||
This years ETHZ Basel project goal is to control E.coli chemotaxis by hijacking and perturbing the tumbling / directed flagellar movement apparatus. By coupling directed flagellar movement regulating proteins to a light-sensitive spatial localization system, their activity can be controlled reversibly. A light-sensitive dimerizing complex fused to this regulating proteins and a spatial fixed location is induced by light pulses and therefore localization of the two molecules can be manipulated. Tumbling / directed flagellar movement rates are supervised by image processing algorithms, which are linked to the light-pulse generator. This system enables to control single E. coli cells to move like mindless "Lemmings" in the direction they are forced to go. | This years ETHZ Basel project goal is to control E.coli chemotaxis by hijacking and perturbing the tumbling / directed flagellar movement apparatus. By coupling directed flagellar movement regulating proteins to a light-sensitive spatial localization system, their activity can be controlled reversibly. A light-sensitive dimerizing complex fused to this regulating proteins and a spatial fixed location is induced by light pulses and therefore localization of the two molecules can be manipulated. Tumbling / directed flagellar movement rates are supervised by image processing algorithms, which are linked to the light-pulse generator. This system enables to control single E. coli cells to move like mindless "Lemmings" in the direction they are forced to go. | ||
---> | ---> | ||
| - | |||
| - | |||
| - | |||
Revision as of 15:24, 5 August 2010
E. lemming
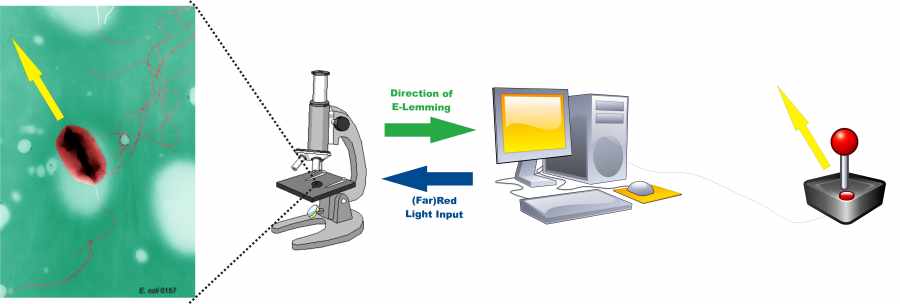
The core idea of our project is to control the movement (chemotaxis) of E. coli- by means of light! In fact, we will alter the chemotactic pathway either by substituting the receptor with a light-sensitive one or by interference with the kinase-phosphatase process. We will realize this with the help of proteins whose association and dissociation can be stimulated by pulses of light waves. In both ways, E. coli tumbling is induced or supressed simply by pressing a light switch! As a consequence, the bacterium can be "driven" to a precise, pre-fixed location. Tumbling / directed flagellar movement rates are monitored by a digital controller, which is combined with image processing algorithms. This system enables control of single E. coli cells to move like mindless "Lemmings" in the direction they are forced to go.
 "
"


