Team:Stockholm/14 July 2010
From 2010.igem.org
m (→Transformation) |
|||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
==Johan== | ==Johan== | ||
| Line 78: | Line 76: | ||
# 72 °C - 5 min | # 72 °C - 5 min | ||
# 4 °C - ∞ | # 4 °C - ∞ | ||
| + | |||
| + | ==Andreas== | ||
| + | |||
| + | ===Clonings=== | ||
| + | ====Colony verifications==== | ||
| + | ''Continued from 13/7'' | ||
| + | |||
| + | =====Restreaks===== | ||
| + | White colonies for all streaked/picked constructs except: | ||
| + | *pSB1K3.SOD (a) | ||
| + | *pSB1C3.SOD (b) | ||
| + | *pSB1K3.BBa_J18931 (a) | ||
| + | *pSB1A3.BBa_J18931 (b) | ||
| + | *pSB1C3.yCCS (a) | ||
| + | |||
| + | =====Gel verifications===== | ||
| + | pEX.X constructs picked and amplified 13/7 were separated on a 1 % gel. No good results, gel results discarded. New gel to be run tomorrow. | ||
| + | |||
| + | ====BL21 ON cultures==== | ||
| + | ''Continued from 13/7'' | ||
| + | |||
| + | BL21 ON cultures of pEX.BBa_J18930-32 were used to inoculate cultures for IPTG induction: | ||
| + | |||
| + | *80 ml LB | ||
| + | *1 % glucose | ||
| + | *100 μg/ml Amp | ||
| + | * 0.8 ml ON culture | ||
| + | Culture grown in 37°C, 250 rpm until an OD<sub>600</sub> of ≈0.5. | ||
Revision as of 17:56, 29 July 2010
Contents |
Johan
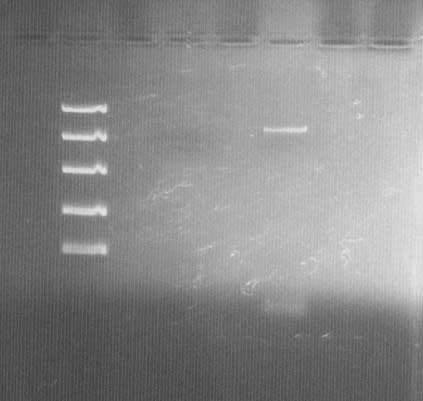
DNA electrophoresis
Lane 1: Fermentas FastRuler™ High Range DNA Ladder ([http://www.fermentas.com/templates/files/tiny_mce/coa_pdf/coa_sm1123.pdf link]), 3: bFGF vector before site-directed mutagenesis PCR and Dpn I treatment (low concentration; 10 ng/µl), 5: bFGF vector after site-directed mutagenesis PCR and Dpn I treatment.
The vector (bFGF-pGEX) shows the correct size of ~5,5k bp. Comparison between lane 3 & 5 also shows that the amplification in the PCR worked and that Dpn I treatment did not cause problems.
Transformation
PCR creates linear DNA, to make the vector circular it has to be transformed; the E. coli makes it circular.
Notes on protocol:
Step 1. Two tubes Bl21 tubes with competent cells were used, one for the linear mutagenesis DNA vector, and one for commercial plasmid as a positive control.
Step 2. 1 µl was used for both transformations.
Step 3. LB was used instead of SOC.
Step 9. No dilutions was made
Step 10. 50 µl was plated. Two plates was used for each reaction, one commercial carbenicillin plate and one of our ampicillin plate (to examine the state of our plates).
Colony PCR
A colony PCR was performed with tubes from Andreas.
(make names more clear..)
- PC 30 A
- PC 31 A
- PC 32 A
- PC 30 B
- PC 31 B
- PC 32 B
- PA 30 A
- PA 31 A
- PA 32 A
- PA 30 B
- PA 31 B
- PA 32 B
- PK 30 A
- PK 31 A
- PK 32 A
- PK 30 B
- PK 31 B
- PK 32 B
- PA SOD A
- PA SOD B
- PC SOD A
- PC SOD B
- PK SOD A
- PK SOD B
- PC yCCS A
- PC yCCS B
- All reaction used VF & VR2 primers. The primers were diluted 15x to get a concentration of 10 µM.
- 33 µm H2O
- 2 µl f.primer 10 µM
- 2 µl r.primer 10 µM
- 1 µl dNTPs 10 mM
- 10 µl Pfu buffer 10x
- 1 µl PfuX7 polymerase
- 1 µl template DNA
- PCR Reaction:
- 98 °C - 2 min
- 31 cycles of
- 98 °C - 10 sec
- 55 °C - 15 sec
- 72 °C - 3 min
- 72 °C - 5 min
- 4 °C - ∞
Andreas
Clonings
Colony verifications
Continued from 13/7
Restreaks
White colonies for all streaked/picked constructs except:
- pSB1K3.SOD (a)
- pSB1C3.SOD (b)
- pSB1K3.BBa_J18931 (a)
- pSB1A3.BBa_J18931 (b)
- pSB1C3.yCCS (a)
Gel verifications
pEX.X constructs picked and amplified 13/7 were separated on a 1 % gel. No good results, gel results discarded. New gel to be run tomorrow.
BL21 ON cultures
Continued from 13/7
BL21 ON cultures of pEX.BBa_J18930-32 were used to inoculate cultures for IPTG induction:
- 80 ml LB
- 1 % glucose
- 100 μg/ml Amp
- 0.8 ml ON culture
Culture grown in 37°C, 250 rpm until an OD600 of ≈0.5.
 "
"