Team:TU Delft/23 July 2010 content
From 2010.igem.org
(Difference between revisions)
(→Alkane Degradation) |
(→Alkane Degradation) |
||
| Line 88: | Line 88: | ||
|0.5 μl KpnI | |0.5 μl KpnI | ||
|A (Boehringer) | |A (Boehringer) | ||
| - | | | + | |✓ |
|271, 2427 | |271, 2427 | ||
|- | |- | ||
| Line 96: | Line 96: | ||
|1 μl NruI | |1 μl NruI | ||
|B (Boehringer) | |B (Boehringer) | ||
| - | | | + | |✓ |
|598, 2109 | |598, 2109 | ||
|- | |- | ||
| Line 104: | Line 104: | ||
|1 μl NruI | |1 μl NruI | ||
|B (Boehringer) | |B (Boehringer) | ||
| - | | | + | |✓ |
|598, 2109 | |598, 2109 | ||
|- | |- | ||
Revision as of 14:28, 27 July 2010
Alkane Degradation
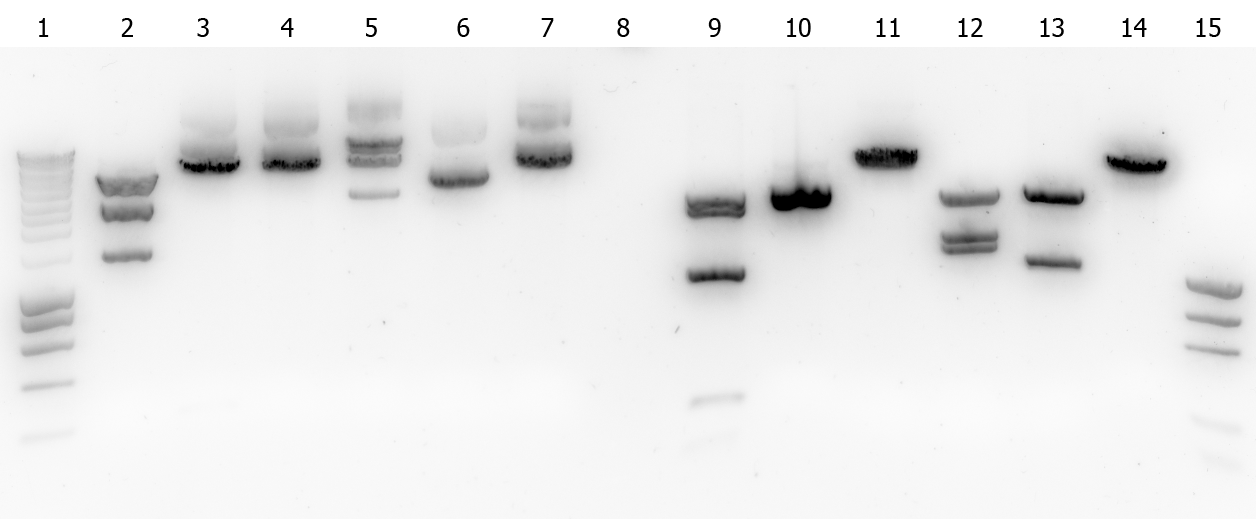
With a number of yesterday's colonies a digestion was done to check if they contain the desired insert. The table below describes the digestion procedure, as well as the gel lane description. (Digestion was done in 10μl end-volume)
| # | Contents | Enzyme 1 | Enzyme 2 | Buffer | BSA | Expected size(s) (bp) |
| 1 | Smartladder (5μl) | n/a | ||||
| 2 | 007T (2) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 1341, 2455 |
| 3 | 008T (4) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 276, 2455 |
| 4 | 009T (6) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 285, 2455 |
| 5 | 009T (7) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 285, 2455 |
| 6 | 010T (9) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 1382, 2455 |
| 7 | 017T (14) | 0.5 μl EcoRI | 0.5 μl PstI | 3 (Biolabs) | ✗ | 855, 2455 |
| 8 | Empty | |||||
| 9 | 007T (2) | 1 μl PstI | 1 μl PvuII | H (Boehringer) | ✗ | 2529,983,251 |
| 10 | 008T (4) | 1 μl AatII | 0.5 μl KpnI | A (Boehringer) | ✓ | 271, 2427 |
| 11 | 009T (6) | 0.5 μl KpnI | 1 μl NruI | B (Boehringer) | ✓ | 598, 2109 |
| 12 | 009T (7) | 0.5 μl KpnI | 1 μl NruI | B (Boehringer) | ✓ | 598, 2109 |
| 13 | 010T (9) | 1 μl EarI | 0.5 μl PstI | 1 (Biolabs) | ✗ | 1075, 2729 |
| 14 | 017T (14) | 0.5 μl AdhI | 0.5 μl SpeI | 4 (Biolabs) | ✗ | 428, 2849 |
| 15 | EZ load (ladder) | n/a |
It seems (from the gel) that BioBrick 007T is the correct one, and the others don't contain the desired insert.
 "
"