Team:Mexico-UNAM-CINVESTAV/Project/Cold shock response
From 2010.igem.org
(→Image:COLDSHOCK.jpg) |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Mexico-UNAM-CINVESTAV-HEADER}} | {{Mexico-UNAM-CINVESTAV-HEADER}} | ||
| - | + | ||
=[[Image:COLDSHOCK.jpg]]= | =[[Image:COLDSHOCK.jpg]]= | ||
[[Image:E_coli.jpg|thumb|150px|Escherichia coli shows a mechanism denominated Cold Shock Response|right]] | [[Image:E_coli.jpg|thumb|150px|Escherichia coli shows a mechanism denominated Cold Shock Response|right]] | ||
| - | All organisms have different mechanisms in order to adapt to variable temperatures. Escherichia coli shows a mechanism denominated Cold Shock Response, which is important since at low temperatures the production of cytoplasmic proteins becomes blocked and E. coli cannot | + | All organisms have different mechanisms in order to adapt to variable temperatures. ''Escherichia coli'' shows a mechanism denominated Cold Shock Response, which is important since at low temperatures the production of cytoplasmic proteins becomes blocked and ''E. coli'' cannot grow (Yamanaka; 1999). |
| - | The CSR consists in | + | The cold shock response (CSR) consists in expressing a specific set of proteins called cold shock proteins (Csp’s) to overcome this translational block. The cold shock protein A (cspA), which is the one we will use, because it is the most studied protein in the family, has tree control processes. The first control point is the auto regulation system Cold Box related to the cold shock protein. The second one is the 5’UTR mRNA region capability to become unstable at 37ºC. The last one is at translation level with the two ribosomes binding sites and an anti-DB sequence in the ribosome which allow to synthesize proteins at 15ºC or a little less (Yamanaka; 1999). A natural question is how the expression of csp’s genes is enabled while the synthesis of most cytoplasm proteins is blocked. First, the cspA mRNA transcription is constitutive both at 37°C and 15°C, but the cold shock proteins expression is only present at low temperatures. This means that the mRNA is synthesized but the expression of proteins depends on the cspA mRNA sequence factors: |
| Line 12: | Line 12: | ||
| - | '''Promoter:''' The promoter consists of 50 bases. This promoter is active at both 37ºC and 15ºC but is more efficient at 15ºC than other promoters because it has an important UP element AT-rich that is | + | '''Promoter:''' The promoter consists of 50 bases. This promoter is active at both 37ºC and 15ºC but is more efficient at 15ºC than other promoters because it has an important UP element AT-rich that is reported to be recognized by the subunit of RNA polymerase and which give it a high promoter activity. |
'''Cold box:''' Is an important region of the 5’ UTR region, which may form a stable stem-loop structure and is implicated in the auto regulation of the cspA system. | '''Cold box:''' Is an important region of the 5’ UTR region, which may form a stable stem-loop structure and is implicated in the auto regulation of the cspA system. | ||
| - | '''5’ UTR:''' Is a region that consists of | + | '''5’ UTR:''' Is a region that consists of essentially 159 bases and is downstream the promoter. This region make the cspA mRNA unstable at 37º C with an average half-life of less than 12 seconds, but more stable at 15°C with an average half-life of 20 minutes and improve the translation at low temperature. Here there is an RNAse cleavage site immediately upstream the Shine-Dalgarno sequence (see below) that is considered to be responsible for this unstable messenger stage. |
| - | + | ||
| - | + | ||
| - | ''' | + | '''Shine-Dalgarno (SD) sequence:''' Is a four-nucleotide sequence that the ribosome uses to bind the mRNA and is common to many messengers also the ones that we use in the IGEM competition. (AAGG) |
| - | ( | + | |
| + | '''DB sequence:''' a fourteen nucleotide sequence that is in the mRNA and complementary to a region found in the 16S rRNA named anti-DB sequence, which provides good translation at low temperature because it functions as an extra messenger ribosome binding site. In this way the ribosome can anchor the SD sequence and the DB sequence as a two ribosome-binding site to permit a great cleavage between the ribosome and the messenger. (Yamanaka; 1999) (Baneyx and Mujacic; 2002) | ||
| - | [[Image: | + | [[Image:Csr2.jpg|thumb|center|550px|left|Cold shock response | Taken from Taken from Horn G, Hofweber R, Kremer W, Kalbitzer HR (2007) Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64: 1457–1470]]Also the cell has many other proteins that are involved in the protein, DNA or RNA stabilization at low temperatures: RecA, which is involved in recombination and induction of the cold shock response; H-NS, a nucleotide-associated DNA-binding protein, which is required for optimal growth at low temperature; GyrA, the subunit of the topoisomerase DNA gyrase, which helps to uncoil the double helix of the DNA; NusA, which is involved in termination of transcription; PNP, which is an exoribonuclease; and Hsc66, a Hsp70 homologue to cold (Thieringer, et al; 1998) (Yamanaka; 1999). |
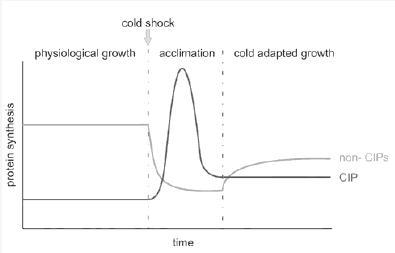
| - | Evidence indicates that the CSIP's express only when the cold shock occurs. Their presence decreases as the growth phase gets closer and | + | Evidence indicates that the CSIP's express only when the cold shock occurs. Their presence decreases as the growth phase gets closer and then is minimal again when the growth phase begins. In the case of non-CSIP's, the expression is normal at physiological temperature. As soon as temperature downshifts the expression decreases and stays low in the acclimation phase. When the growth phase starts the non-CSIP's increase their expression but stay at a low rate (Horn et al.; 2007) |
Latest revision as of 03:55, 28 October 2010
All organisms have different mechanisms in order to adapt to variable temperatures. Escherichia coli shows a mechanism denominated Cold Shock Response, which is important since at low temperatures the production of cytoplasmic proteins becomes blocked and E. coli cannot grow (Yamanaka; 1999).
The cold shock response (CSR) consists in expressing a specific set of proteins called cold shock proteins (Csp’s) to overcome this translational block. The cold shock protein A (cspA), which is the one we will use, because it is the most studied protein in the family, has tree control processes. The first control point is the auto regulation system Cold Box related to the cold shock protein. The second one is the 5’UTR mRNA region capability to become unstable at 37ºC. The last one is at translation level with the two ribosomes binding sites and an anti-DB sequence in the ribosome which allow to synthesize proteins at 15ºC or a little less (Yamanaka; 1999). A natural question is how the expression of csp’s genes is enabled while the synthesis of most cytoplasm proteins is blocked. First, the cspA mRNA transcription is constitutive both at 37°C and 15°C, but the cold shock proteins expression is only present at low temperatures. This means that the mRNA is synthesized but the expression of proteins depends on the cspA mRNA sequence factors:
Promoter- cold box - 5’UTR - SD -DB
Promoter: The promoter consists of 50 bases. This promoter is active at both 37ºC and 15ºC but is more efficient at 15ºC than other promoters because it has an important UP element AT-rich that is reported to be recognized by the subunit of RNA polymerase and which give it a high promoter activity.
Cold box: Is an important region of the 5’ UTR region, which may form a stable stem-loop structure and is implicated in the auto regulation of the cspA system.
5’ UTR: Is a region that consists of essentially 159 bases and is downstream the promoter. This region make the cspA mRNA unstable at 37º C with an average half-life of less than 12 seconds, but more stable at 15°C with an average half-life of 20 minutes and improve the translation at low temperature. Here there is an RNAse cleavage site immediately upstream the Shine-Dalgarno sequence (see below) that is considered to be responsible for this unstable messenger stage.
Shine-Dalgarno (SD) sequence: Is a four-nucleotide sequence that the ribosome uses to bind the mRNA and is common to many messengers also the ones that we use in the IGEM competition. (AAGG)
DB sequence: a fourteen nucleotide sequence that is in the mRNA and complementary to a region found in the 16S rRNA named anti-DB sequence, which provides good translation at low temperature because it functions as an extra messenger ribosome binding site. In this way the ribosome can anchor the SD sequence and the DB sequence as a two ribosome-binding site to permit a great cleavage between the ribosome and the messenger. (Yamanaka; 1999) (Baneyx and Mujacic; 2002)
Evidence indicates that the CSIP's express only when the cold shock occurs. Their presence decreases as the growth phase gets closer and then is minimal again when the growth phase begins. In the case of non-CSIP's, the expression is normal at physiological temperature. As soon as temperature downshifts the expression decreases and stays low in the acclimation phase. When the growth phase starts the non-CSIP's increase their expression but stay at a low rate (Horn et al.; 2007)
Baneyx, François and Mujacic, Mirna; (2002); Cold-Inducible Promoters for Heterologous Protein Expression; from Methods in Molecular Biology; Vol. 205 Humana Press Inc.[http://www.springerlink.com/content/qx66q8875x0q7043/#section=86530&page=1]
Horn G., et al.; (2007); Structure and function of bacterial cold shock proteins; Cellular and Molecular Life Sciences; Vol. 64 p. 1457 – 1470
Thieringer A. Heather, Pamela G. Jones, and Masayori Inouye; (1998); Cold Shock and Adaptation; Bio Essays 20.1: 40-57[http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1521-1878(199801)20:1%3C49::AID-BIES8%3E3.0.CO;2-N/abstract]
Yamanaka, Kunitoshi; (1999); Cold Shock Response in Escherichia coli; Journal of Molecular and Microbiological Biotechnology 1(2): 193-202[http://www.horizonpress.com/jmmb/v1/v1n2/03.pdf]
 |
 |
 |
 |
 "
"