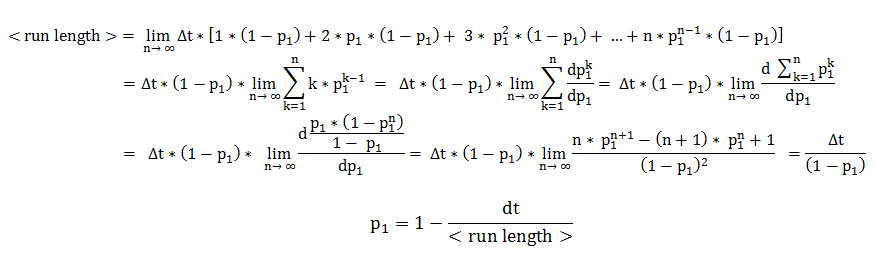Team:ETHZ Basel/Modeling/Movement
From 2010.igem.org
(→Overview) |
|||
| Line 3: | Line 3: | ||
= Modeling & Simulating Bacterial Movement = | = Modeling & Simulating Bacterial Movement = | ||
| - | |||
== Overview == | == Overview == | ||
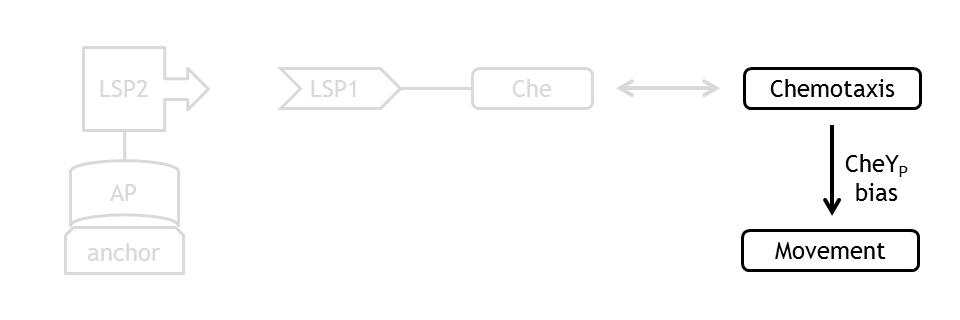
| + | [[Image:ETHZ_Basel_molecular_move.png|thumb|400px|'''Schematical overview of the relation between the chemotaxis pathway and the movement model.''']] | ||
Analogous to the highly complex signal transduction network in the chemotaxis receptor model, there is an at least equally challenging molecular mechanism on side of the flagella, responsible for the movement of the bacterium. | Analogous to the highly complex signal transduction network in the chemotaxis receptor model, there is an at least equally challenging molecular mechanism on side of the flagella, responsible for the movement of the bacterium. | ||
Revision as of 02:07, 28 October 2010
Modeling & Simulating Bacterial Movement
Overview
Analogous to the highly complex signal transduction network in the chemotaxis receptor model, there is an at least equally challenging molecular mechanism on side of the flagella, responsible for the movement of the bacterium.
In order to capture the features of cell motility, we not only implemented, but also conceptually created a probabilistic two state model, which simulates the cell movement in accordance to the existing empirical observations.
At every time point of the simulation of the movement model, the current CheYp concentration is received as an input from the chemotaxis pathway model. This input determines the probability of the CCW and the CW rotation direction of the flagella. A counterclockwise rotation corresponds to the directed movement (running) state and to a change in spatial coordinates, together with a slight change in angle, while a clockwise rotation corresponds to the tumbling state and to a new movement direction, with nearly unchanged spatial coordinates.
By coupling the probabilistic movement model with the deterministic molecular model we can simulate the variations in motility which occur due to variations in the concentrations of the molecular species of the chemotaxis pathway, namely in the concentration of CheYp.
Known Features of Chemotaxis Motility
The focus in creating the movement model was to reliably reproduce the known chemotaxis behavior of E. coli cells. We used as a benchmark the statistical estimates of chemotaxis motility, as presented in [1]. In order to be successful, our movement model had to reproduce the following features:
- In a constant extracellular environment, the transition times between the two states the cell can employ (run length and tumbling length) are approximately exponentially distributed, with a tumbling length of 0.14±0.19s and a run length of 0.86±1.18s (each value given as mean and standard deviation).
- In a changing extracellular environment (e.g. increasing concentration of Aspartate), mainly the distribution of the run length adjusts, whereas the distribution of the tumbling length only slightly changes.
- The average speed of an E. coli during directed movement is approximately 14.2±3.4μm/s, whereas during tumbling the average speed is significantly smaller.
- In a constant extracellular environment, the absolute angle change between two directed movement phases is Weibull distributed, with a non-zero maximum and a mean and standard deviation of 68±36°, whereas the absolute angle change during one directed movement period has its maximum around 0°, with mean and standard deviation 23±23°.
Assumptions
Mean Times
In accordance with the experimental data given in [1], the mean tumbling length was assumed to be independent of the extracellular conditions, unlike the mean run length, which is known to fluctuate for wild type cells under different extracellular environments. Besides the experimental evidence in [1], this assumption also has a biological interpretation: tumbling is believed to occur because the flagella push each other away and disassemble after rotating counter-clock wise (directed movement). The time spent in tumbling is a stochastic variable mainly representing the time the flagella need to reassemble afterwards, and a single process of reassembling is only weakly correlated with the frequency of tumbling.
Velocity
During the directed movement phase, the speed of an E. coli cell varies only slightly around its mean value, whereas during the tumbling phase, the speed is significantly smaller and can be neglected (see [1]).
Therefore, we assumed in our model that the velocity during directed movements is constant and zero during tumbling.
Direction Changes
During one directed movement phase, the angular speed is Gaussian distributed, with zero mean and a tuned standard distribution, such that, when integrated over the mean run length, we obtain the properties stated above and in [1].
Reproducing the distribution for the angular change between two directed movement phases was much more challenging. Choosing the angular speed independently at every integration step, as we did for the directed movement case, would lead to a distribution which reaches its maximum at zero, which would be in discrepancy with the empirical data in [1]. Therefore, we decided to assume a constant angular speed for the entire period between two directed movement phases. The angular speed was drawn from a tuned distribution, which was optimized to reflect the observed data as good as possible.
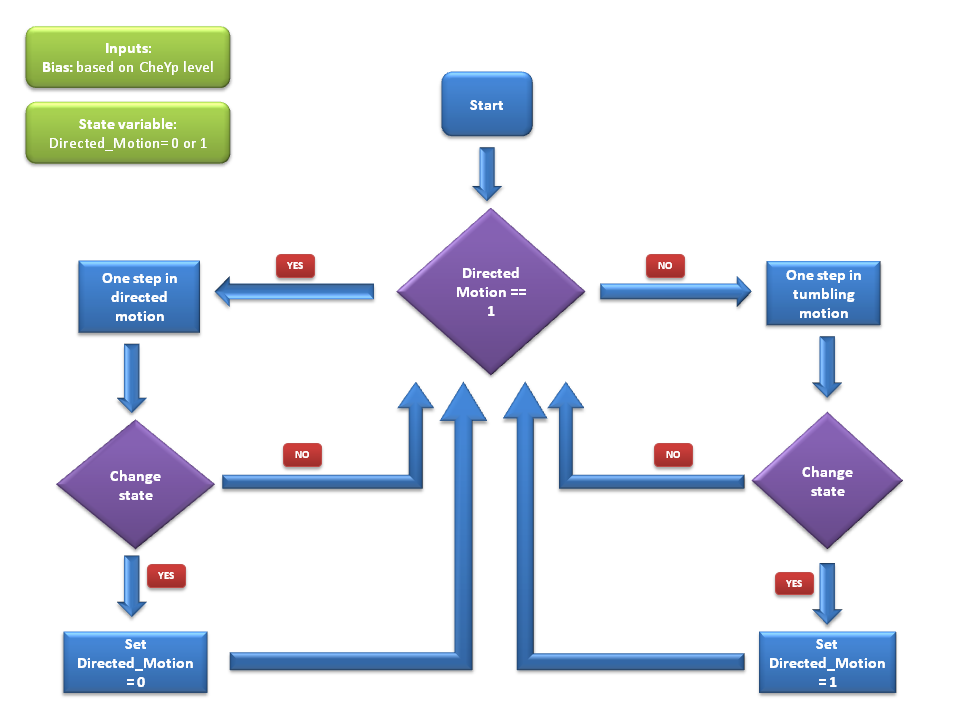
General Algorithm
We use a two state model to simulate bacterial motility, with the two states corresponding to directed movement and to tumbling. At each time step of the simulation, the cell either keeps or switches its state, according to the input received from the molecular model, which determine the values of the transition probabilities between states.
If the model is currently in the directed movement state, the cell mainly moves in its current direction with a speed of 14.2μm/s and changes direction only slightly (see [1]).
If however the model is currently in the tumbling state, the cell is changing its direction rapidly while staying at the same point.
Coupling to the Chemotaxis Pathway
The quantity that links the CheYp concentration with the type of motion (tumbling vs. directed motion) is called the bias and it is formally defined as the fraction of time spent in the directed movement state with respect to the total time in motion. Our choice for bias formulation was a nonlinear Hill - type function of CheYp concentration, with [CheYp]wt being the wild-type CheYp concentration for which a steady - state invariant time ratio was obtained. Both the functional dependency and CheYp value are well documented in the literature [2].
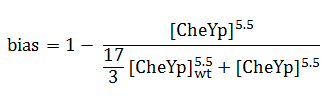
Since the mean tumbling length is assumed to be independent of the intracellular state, the mean run length has to be a function of the bias:

Transition Probabilities
At every time step of the simulation, the model decides probabilistically whether to switch or to keep its current movement state. Therefore, bacterial motility can be modeled as a two - state first - order Markov process, in which the future state is only dependent on the current state. Since we have Boolean values for both directed movement and tumbling, we can define the following four transition probabilities, separately controlled depending on whether the cell is currently in the directive movement or in the tumbling phase.
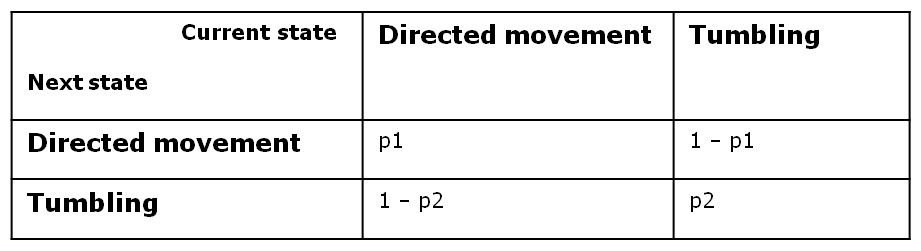
Therefore, the two central parameters of our model are the probabilities of keeping the current state also as future state: p1: the probability of the future state being directed movement, when the current state is directed movement and p2: the probability of the future state being tumbling, when the current state is tumbling.
In deriving the expression of these probabilities, we separately and symmetrically focused on the two processes: directed movement and tumbling. As a consequence, the mean run length is only dependent on the time step and on the probability of continuing the directed movement (p1), while the mean tumbling length is only dependent on the time step and on the probability of continuing tumbling (p2).
We will explain in detail the technique employed in deriving the probability of being in the directed movement state, when the previous state was directed movement. Symmetrical calculations, for the mean tumbling length, follow identically.
The mean run length is the expected value of a random variable representing the number of time steps the cell consecutively spends in directed movement. By expanding the definition of an expected value, the mean run length becomes an infinite sum over all possible consecutive run-lengths, multiplied by their respective occurrence probabilities.
Direction Changes
Angular Velocity during Tumbling
As stated above, the observed probability distribution of the absolute angle change between two directed movement phases is not maximal around zero. An intuitive approach, which would set the angular speed to a randomly positive or negative value drawn independently from a symmetric probability distribution, would always result in a distribution having its maximum around zero, therefore not reproducing the real motility behavior.
We thus decided to keep the angular velocity fixed for an entire tumbling phase. However, even with this approach, it was not possible to fully reproduce the quantitative properties of the observed distribution.
The reason for this discrepancy was that the shape of the distribution of the tumbling lengths makes it impossible to create a distribution of the angle changes corresponding in all of its properties to the observed one, without making the angular speed inversely proportional to the tumbling length. Although such an approach is easy to implement, the resulting model would be characterized by a lack of mathematical aesthetic.
Therefore, the distribution was chosen to be independent on the tumbling length and tuned to as good as possible fit the empirical one (see [1]). The resulting distribution had, as the observed one, a nonzero maximum, and a mean between 60° and 70°. However, the standard deviation was 47°, 11° higher than the observed one of 36°.
Angular Velocity during Directed Movement
Different to the angular velocity during tumbling, the angular velocity during directed movement phases can be drawn from a tuned distribution to fully reproduce the experimental data (see [1]): mean of 23° and standard deviation of also 23°. We assumed that the angular speed during one time step is normally distributed with zero mean and a standard deviation of σ. Thus, as one directed movement period consists of N=<run length>/Δt integration steps, the change in angle after one directed movement period has zero mean and a standard deviation of
σrun=σ Δt N½ = σ<run length>½Δt½.
The literature values of change in angle are however given as absolute values. This corresponds to calculating χ-distribution of the angular change, which has in this case the mean value of μχ=(2/π)½σrun and a standard deviation of σχ=σrun. Since (2/π)½≈0.8 and thus the resulting mean and standard deviations are similar to each other as experimentally observed, we have chosen a standard deviation of
σ=(π/2)23°<run length>-½Δt-½for our model. The resulting distribution of the angular change during directed movement periods resembles closely to the experimentally available values (see simulation results).
Simulation Results
For our simulations, we have chosen a timestep Δt=0.03s, which is around five times smaller than the smallest time constants in the cell motility model, as well as over ten times smaller than the time constant with which the CheYp concentration changes in the molecular model (for details, see optimal experimental design).
Evaluation of the Model
To evaluate the movement model, we performed several long time simulations under constant conditions (bias set to its wild-type steady state value of around 0.85) and determined its characteristics which might be compared to the known properties of the motility of E. coli cells as given in [1] and described above.
 Distribution of the absolute angle change between two directed movement phases. The distribution has similar properties as the one experimentally observed in [1]. Its mean value is 62.3° and its standard deviation 47.0°. |
Variations in input bias
Furthermore, we simulated the spatial behavior of a single E. coli cell under the influence of different input bias values. For a more realistic simulation of the bacterial movement process, the input bias was obtained directly from the combined molecular model linking the chemotaxis pathway and the light switch. As the combined model offered us many design alternatives for performing simulations, we decided on a fixed molecular template, to ease the comparison of results.
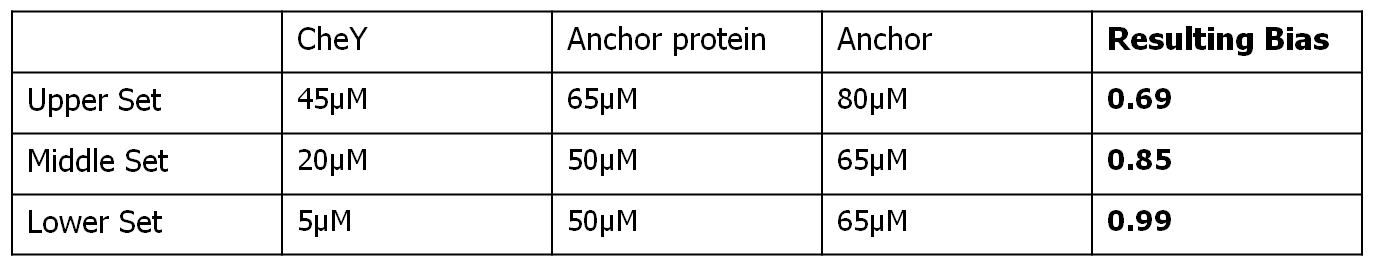
The chosen molecular template for simulating the movement model was the following:
- molecular model chosen: Spiro et al. [3]
- controlled Che species: CheY
- Light coupling: CheY-PIF3 & PhyB-Anchor
Each row of the subplots table presented below represents a different input bias value for the movement model, obtained as a result of different input protein concentrations for the combined molecular model. Each column represent a different simulation length (100 seconds, 500 seconds and 3000 seconds).
The input protein concentrations used, together with the corresponding obtained steady-state bias were the following:
All the simulations were run without the existence of any controller algorithm, therefore reproducing the random chemotaxis walk.
Please note the different spatial ranges when comparing the plots, increasing both with increasing simulation time length (the bacterium travels a longer distance in a longer time period) and with increasing bias value (the probability of choosing the directed movement state increases, therefore, on average, the cell travels a longer distance).
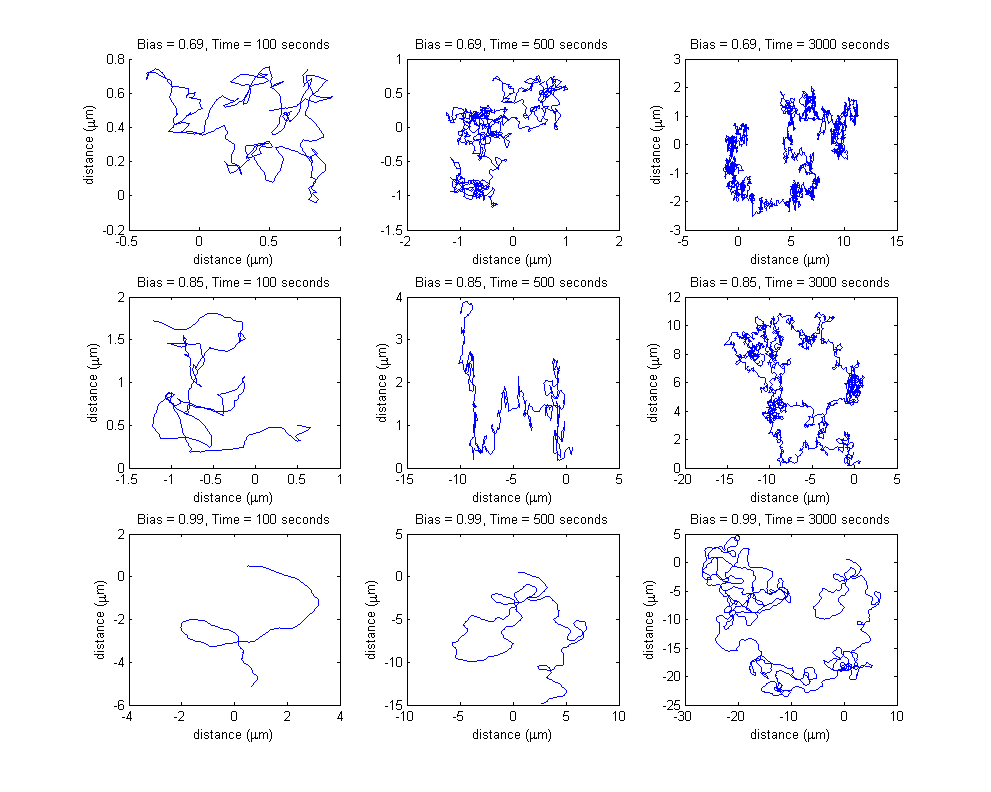
Note the increasing axis range, both on the x and on the y coordinate, due to both increasing bias value and increasing simulation time.
Directed movement/Tumbling
For the above simulated trajectories, we generated the so-called directed movement/tumbling plots used in analyzing chemotaxis motility from the perspective of boolean movement - time dependency. For providing a better visibility of the individual steps of the movement process, in the left figure only the first 60 seconds of the above simulations were considered, while in the right figure a detailed insight into the first 30 seconds is provided.
Download
The bacterial movement model is included within the Matlab Toolbox and can be downloaded there.
References
[1] [http://www.nature.com/nature/journal/v239/n5374/abs/239500a0.html H.C. Berg, D.A.Brown: Chemotaxis in Escherichia coli analysed by three - dimensional Tracking. Nature 1972 239, 500 - 504]
[2] [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1299372/?tool=pubmed Levin, Morton-Firth: Origins of Individual Swimming Behavior in Bacteria, Biophysical Journal 1998 74:175-181]
[3] [http://www.pnas.org/content/94/14/7263.full Spiro et al: A model of excitation and adaptation in bacterial chemotaxis. PNAS 1997 94;14;7263-7268.]
 "
"