Team:Slovenia/PROJECT/proof/domain
From 2010.igem.org
| (2 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<style> | <style> | ||
#vsebina_mid{ | #vsebina_mid{ | ||
| - | height: | + | height: 5400px; |
} | } | ||
#lgumb2{ | #lgumb2{ | ||
| Line 31: | Line 31: | ||
<h2>TAL effectors</h2> | <h2>TAL effectors</h2> | ||
| - | TAL family of proteins consists of transcription activator like effectors from plant pathogenic bacteria of Xanthomonas spp. TAL effectors are proteins that are secreted into eukaryotic cells by type III secretion system. In host, they are translocated to the nucleus where they modulate transcription of different genes. TAL protein consists of signal N-terminal, C-terminal domain with transcription activator activity and central region of tandem repeats. It was shown that hypervariable pair of amino acids in every tandem repeat of TAL protein defines its binding properties (Boch et al., 2009). As such TAL proteins can be programed to bind any desired nucleotide sequence. | + | TAL family of proteins consists of transcription activator like effectors from plant pathogenic bacteria of ''Xanthomonas spp.'' TAL effectors are proteins that are secreted into eukaryotic cells by type III secretion system. In host, they are translocated to the nucleus where they modulate transcription of different genes. TAL protein consists of signal N-terminal, C-terminal domain with transcription activator activity and central region of tandem repeats. It was shown that hypervariable pair of amino acids in every tandem repeat of TAL protein defines its binding properties (Boch et al., 2009). As such TAL proteins can be programed to bind any desired nucleotide sequence. |
Our aim was to design synthetic TAL DNA-binding proteins and use them in our project. As described in literature, designing of TAL proteins with specificity to any given nucleotide sequence should be an easy task (Boch et al., 2009). Central region of TAL protein generally consists of 10 – 18 highly similar tandem repeats. Each repeat is 34 amino acids long. Almost all but two hypervariable amino acids at positions 12 and 13, between different repeats and different TAL effectors are conserved. As reported, these two amino acids define the binding properties of a single tandem repeat. Tandem repeats are interchangeable in nature, and can be shuffled as needed. What is most important, the code for recognition of specific nucleotide in DNA sequence by hypervariable pairs of amino acids is known. Because of modular nature and known recognition code TAL proteins seem to be an ideal platform for designing novel specific DNA-binding proteins with uniform properties. | Our aim was to design synthetic TAL DNA-binding proteins and use them in our project. As described in literature, designing of TAL proteins with specificity to any given nucleotide sequence should be an easy task (Boch et al., 2009). Central region of TAL protein generally consists of 10 – 18 highly similar tandem repeats. Each repeat is 34 amino acids long. Almost all but two hypervariable amino acids at positions 12 and 13, between different repeats and different TAL effectors are conserved. As reported, these two amino acids define the binding properties of a single tandem repeat. Tandem repeats are interchangeable in nature, and can be shuffled as needed. What is most important, the code for recognition of specific nucleotide in DNA sequence by hypervariable pairs of amino acids is known. Because of modular nature and known recognition code TAL proteins seem to be an ideal platform for designing novel specific DNA-binding proteins with uniform properties. | ||
| Line 94: | Line 94: | ||
<hr> | <hr> | ||
| - | Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A. and Bonas U. 2009. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science, 326: 1509-1512. | + | Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A. and Bonas U. 2009. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science, 326: 1509-1512. <br><br> |
| + | Sera T. 2009. Zinc-finger-based artificial transcription factors and their applications. Advanced Drug Delivery Reviews, 61, 7-8: 513–526 | ||
<!--STOP BESEDILO--> | <!--STOP BESEDILO--> | ||
Latest revision as of 01:47, 28 October 2010
Contents |
DNA binding domains - which one to choose?
Arrangement of different functional proteins on DNA molecule is required for implementation of our ideas. Because enzymes and other functional polypeptides usually do not have specific affinity for DNA molecule, we decided to fuse them with specific DNA binding domains. These DNA binding domains should have strong affinity for DNA molecule, but interaction should also be sequence specific. Several classes of DNA binding proteins exist in nature, e.g. zinc finger family of proteins, TAL effectors, different transcription factors (TetR, CI, Gal4, ToxR, CinR, RhlR, LuxR) and endo/exonucleases with wrecked cutting activity. Unfortunately, most of them proved to be unusable due to multimerization nature, low affinity and low sequence specificity. Zinc fingers and TAL effectors were on the top of our list since the beginning, because they are modular in nature and can be programed into practically unlimited number of different sequence specific DNA binding domains.
TAL effectors
TAL family of proteins consists of transcription activator like effectors from plant pathogenic bacteria of Xanthomonas spp. TAL effectors are proteins that are secreted into eukaryotic cells by type III secretion system. In host, they are translocated to the nucleus where they modulate transcription of different genes. TAL protein consists of signal N-terminal, C-terminal domain with transcription activator activity and central region of tandem repeats. It was shown that hypervariable pair of amino acids in every tandem repeat of TAL protein defines its binding properties (Boch et al., 2009). As such TAL proteins can be programed to bind any desired nucleotide sequence.
Our aim was to design synthetic TAL DNA-binding proteins and use them in our project. As described in literature, designing of TAL proteins with specificity to any given nucleotide sequence should be an easy task (Boch et al., 2009). Central region of TAL protein generally consists of 10 – 18 highly similar tandem repeats. Each repeat is 34 amino acids long. Almost all but two hypervariable amino acids at positions 12 and 13, between different repeats and different TAL effectors are conserved. As reported, these two amino acids define the binding properties of a single tandem repeat. Tandem repeats are interchangeable in nature, and can be shuffled as needed. What is most important, the code for recognition of specific nucleotide in DNA sequence by hypervariable pairs of amino acids is known. Because of modular nature and known recognition code TAL proteins seem to be an ideal platform for designing novel specific DNA-binding proteins with uniform properties.
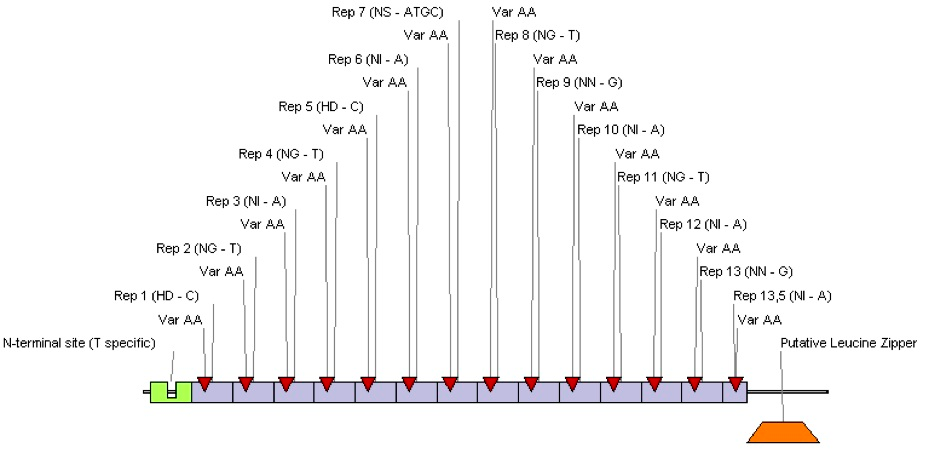
We designed TAL protein with the same predicted binding sequence as Tet operator. For purpose of our project only central DNA-binding domain of TAL protein was preserved, but parts of N-terminal, and C-terminal regions were eliminated, since they have been shown to mediate signaling and transcriptional effects in plants. This modified nucleotide sequence was assembled on a computer and the actual synthetic gene was ordered at GeneArt company. We confirmed specific binding of our novel TAL protein to Tet operator in vivo, using our universal device. At the end we decided not to choose TAL effectors as DNA binding domains for our project, mainly because designing TAL proteins is still in its early stage.
Zinc fingers
At the end we chose zinc finger proteins as DNA binding domains for our project. Zinc finger proteins form a small, independently folded zinc containing mini domain that recognizes specific nucleic acid sequence. They are present in eukaryotic cells as transcriptional factors and are involved in regulation of endogenous gene expression by specific DNA binding. However, some zinc finger proteins are able to bind to RNA or proteins as well. It is estimated that human genome encodes for more than 700 Cys2His2 zinc finger coding genes, thus being the most common DNA-binding proteins in eukaryotes. Nevertheless, recent biochemical and phylogenetical studies confirmed that they aren't restricted only to eukaryota but are also present in prokaryotes.
Biochemical and binding properties of zinc fingers
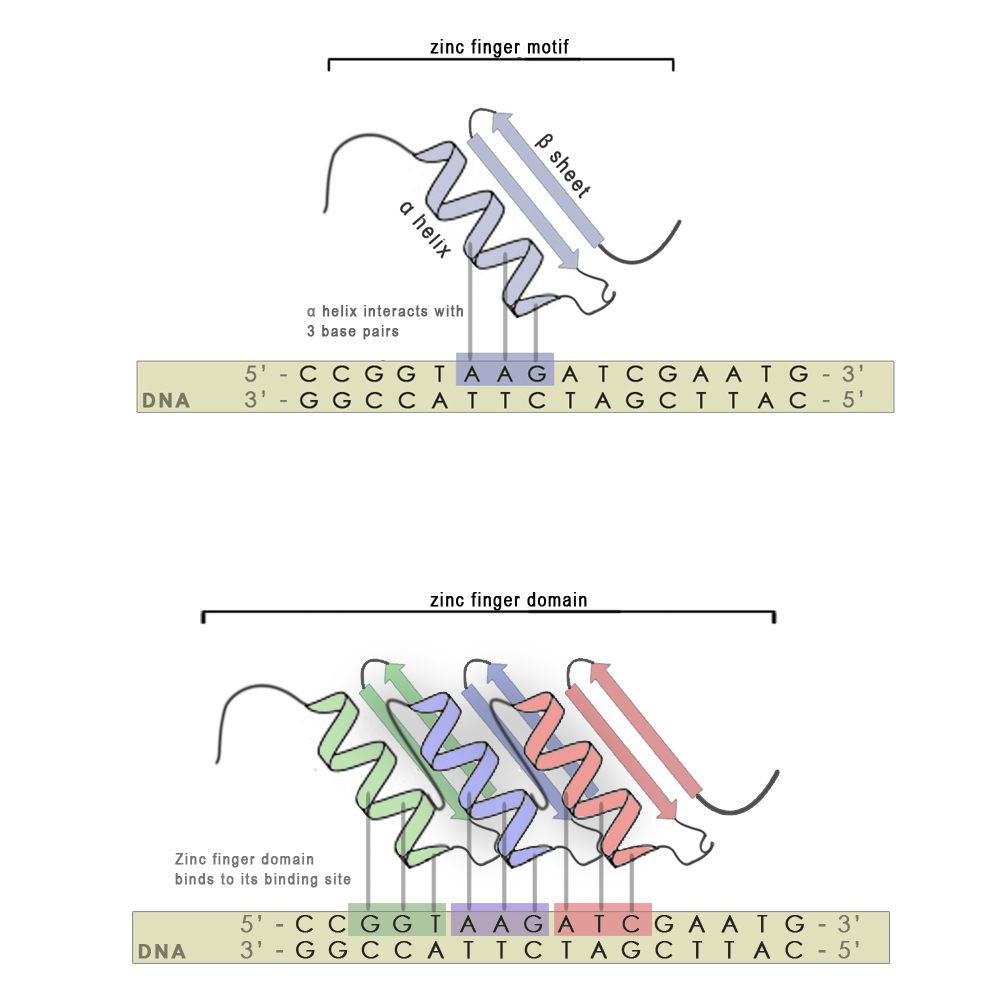
Zinc finger motifs are formed from a short stretch of amino acid residues folded around a zinc cation and have the shape of a finger. Each individual finger is highly conserved in its structure and consists of around 30 amino acid residues. There are two hisitdine residues in every finger on the α-helix and two cysteine residues on the β-sheet that interact with the zinc atom. A short stretch of amino acid residues in the α-helix is responsible for 3 - 4 bp binding specificity. Each finger recognizes and binds to 3 base-pair subsites (see scheme, upper panel), however using modular approach we can stitch several zinc finger mini domains together (scheme, bottom panel) and achieve chemical distinctiveness through variations in certain key amino acid residues. With every new attached zinc finger domain 3-4 base pairs are added to the recognized sequence, thus minimizing non-specific binding on nucleic acids present in host organisms and increasing the specificity of the zinc finger. They also have an advantage over most other DNA binding motifs from natural transcription factors, since the targeted DNA sequence does not have to be symmetric.
Application of zinc finger proteins
In 1994 it was demonstrated for the first time that zinc finger proteins could modulate endogenous gene expression in living cells. Ever since, many studies have been made where promising results were shown using zinc fingers as endogenous gene regulators. They can bind with nanomolar dissociation constants and discriminate effectively between specific and non-specific DNA sites and thus function as transcriptional activators or repressors in living cells. Different groups showed the importance of zinc fingers in regulating gene expression like Erb-2 gene involved in the development of human malignancies, MDR1 gene which is important in controlling multidrug resistance in cancer cells, activation of the human erythropoietin gene and vascular endothelial growth factor A (VEGF-A) and many other genes. It was also shown that zinc fingers could have an important role in antiviral therapies, for example against human immunodeficiency virus type 1 (HIV-1), herpes simplex virus 1(HSV-1) and human papillomavirus (HPV) (Sera, 2009).
The role of zinc finger domains in our project
In this project, we used zinc finger proteins as adaptor molecules that recognize and bind specific DNA sequences instead of their original function as transcription factors. Every selected zinc finger was linked to functional polypeptide that had to retain its biological function. The advantages of using zinc finger domains in our project are their specificity of binding to DNA sequences and the possibility to combine different zinc fingers together, to even further increase their specificity. Our DNA program sequence was designed to organize zinc finger domains linked to functional proteins (split GFPs or biosynthetic enzymes) into a functional complex. We were able to show that functional proteins linked to zinc finger domains bind to the specific target sites on a DNA program, and that biosynthetic enzymes in such ordered state improve the yield of biosynthetic pathway.
Choosing zinc finger domains
First of all we searched through Registry of standard biological parts for existing zinc finger proteins. We found three poorly characterized parts: [http://partsregistry.org/Part:BBa_K165007 BBa_K165007] (Gli-1 DNA binding domain), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165006 BBa_K165006] (Zif268-HIV DNA-binding domain) and [http://partsregistry.org/Part:BBa_K165008 BBa_K165008] (YY1 DNA-binding domain). We compared information about these three zinc fingers found on Registry with information from literature. We decided to use Gli-1 zinc finger and Zif268-HIV zinc finger. The latter was renamed to Zfn HIVC, based on amino acid sequence comparisons and information about the DNA binding site found in literature. We did not use zinc finger YY1 due to its four finger structure. Additional information about Gli-1 and ZnfHIVC that was gathered from the literature and acquired in our experimental work was re-entered to the Registry.
In order to realize our project goal at least 5 more DNA binding domains were needed. We chose to limit our search to zinc fingers with 3 fingers and binding length of 9-10 nucleotides, that have have known binding affinity and that the affinity for similar sequences was very low. Our literature search resulted in 13 putative zinc fingers that met our requirements. So the question was how to choose 7 additional zinc fingers out of 13. We decided to write a computer software for that pourpose and named it ZNF Buster.
ZNF Buster returned the following results that best matched our criteria.
- [http://partsregistry.org/Part:BBa_K165007 Gli-1]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165006 ZNF HIVC]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323105 Zif 268]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323107 PBSII]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323106 Tyr456]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323109 Blues]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323108 Jazz]
ZNF Buster
When choosing 7 zinc fingers out of 13 we wanted to avoid picking a group of zinc finger proteins that bind to similar DNA sequences. Programs that are able to find these similarities already exist, however, they can only compare one zinc finger versus all others individually. To take things a step further, we decided to write a program that will help us find out if any new similarities arise when we combine more zinc fingers with a spacer of 2 base pairs between each pair. The problem of finding the right subgroup turned to be too difficult even for a computer due to the astronomical number of possible combinations.
To make things easier we decided to only test each zinc finger against every other individually, and against all possible pairs with a 2 base pair spacer. That way sufficient information for picking a sub-optimal group of zinc fingers for our needs could be provided. Furthermore, the problem would be simple enough for a computer to solve it in a timely manner.
The program consists of three different tests. First test compares each zinc finger to all others individually, while the second test compares each zinc finger to all possible pairs. The second test consists of two parts - the first part compares the zinc finger to all pairs, catching only same sequences of nucleotides (we set the size of this sequence), while the other part compares all the nucleotides and saves the highest numbers of same nucleotids even if they're not in a sequence. The third test then picks the most suitable sub-group for each zinc finger and calculates its 'score'. The score of the group depends on three parameters the user chooses before starting the program - these three parameters determine the most important test when calculating the score. First parameter increases the value of individual compares to other zinc fingers, the second parameter increases the value of comparing zinc finger to pairs using sequences and the third parameter increases the value of comparing zinc fingers to pairs without sequences.
To follow the BioBrick standard we also made sure that zinc finger or pairs with spacers do not contain any BioBrick restriction sites.
Once these tests are completed, the program takes five groups with the best score, sorts them and prints them. Output print contains the names of the zinc fingers and the best possible 2 base pair spacer for all zinc finger pairs in the group. The following snapshot shows how the output looks like the following figure:
Because this program was initially written to find the best possible sub-group for our project, it is yet possible to test any available zinc finger. In addition, its source code is quite hard to understand and its usage limited to those skilled in using linux shell and java (most of the input is hardcoded, meaning that input is a part of code). However, we figured that others might need a similar tool, so we decided to rewrite this program after we return from MIT and make it user friendly and accessible to wider range of people. It is most likely going to take a form of an applet on our iGEM wiki page once it unlocks or on the webpage of our National Institute of Chemistry.
Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A. and Bonas U. 2009. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science, 326: 1509-1512.
Sera T. 2009. Zinc-finger-based artificial transcription factors and their applications. Advanced Drug Delivery Reviews, 61, 7-8: 513–526
 "
"