Results usu.html
From 2010.igem.org
(→Gold Medal) |
|||
| (4 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
=Synechocystis Promoter Expression in ''E. coli''= | =Synechocystis Promoter Expression in ''E. coli''= | ||
| - | Promoter regions, 5’ Untranslated regions (5’UTR), and ribosome binding sites (RBS) from 11 ''Synechocystis'' genes were cloned upstream of GFP. While these parts are eventually going to be characterized and used in ''Synechocystis'', since the final integration plasmid was not complete, we decided to test these parts in ''E. coli''. We believed that these parts would function in ''E. coli'' due to the similarity of the sigma factor binding sites and RBS sequences from ''Synechocystis'' to the consensus promoter and RBS in ''E. coli'' (Tables | + | Promoter regions, 5’ Untranslated regions (5’UTR), and ribosome binding sites (RBS) from 11 ''Synechocystis'' genes were cloned upstream of GFP (Table 1). While these parts are eventually going to be characterized and used in ''Synechocystis'', since the final integration plasmid was not complete, we decided to test these parts in ''E. coli''. We believed that these parts would function in ''E. coli'' due to the similarity of the sigma factor binding sites and RBS sequences from ''Synechocystis'' to the consensus promoter and RBS in ''E. coli'' (Tables 2 and 3). |
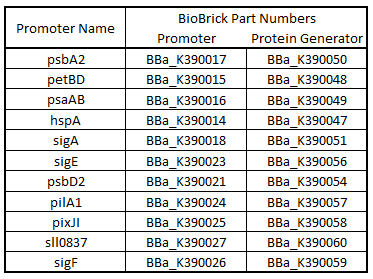
| + | '''Table 1.'''Table of BioBrick Part Numbers and Promoters. | ||
| - | '''Table | + | [[Image:USU_Part_Promoter_Names.png]] |
| + | |||
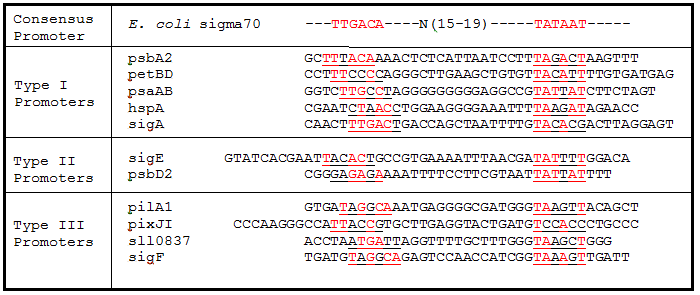
| + | '''Table 2.''' Sigma factor binding site alignments. Underlined sequences are the binding region for sigma70 sigma factor from ''E. coli''. Bases in red are exact matches to the consensus sequence. | ||
[[Image:USU_Promoter_Alignments.png]] | [[Image:USU_Promoter_Alignments.png]] | ||
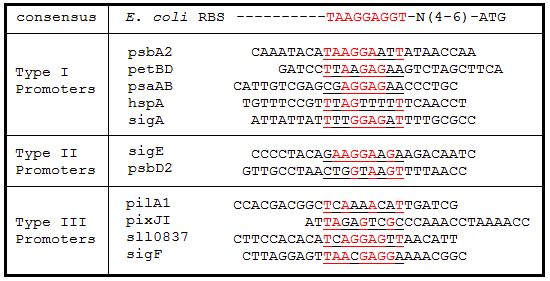
| - | '''Table | + | '''Table 3.''' Ribosome binding site sequence alignments. The regions 25 bp upstream of the start codon for each gene were aligned to the ''E. coli'' consensus RBS sequence. Underlined sequences are the binding region for the 9 bp 3’ end of the 16S rRNA subunit in ''E. coli''. Bases in red are exact matches to the consensus sequence. |
[[Image:USU_RBS_Alignment.png]] | [[Image:USU_RBS_Alignment.png]] | ||
| Line 38: | Line 41: | ||
[[Image:USU_Exponential_Phase.JPG||600px]] | [[Image:USU_Exponential_Phase.JPG||600px]] | ||
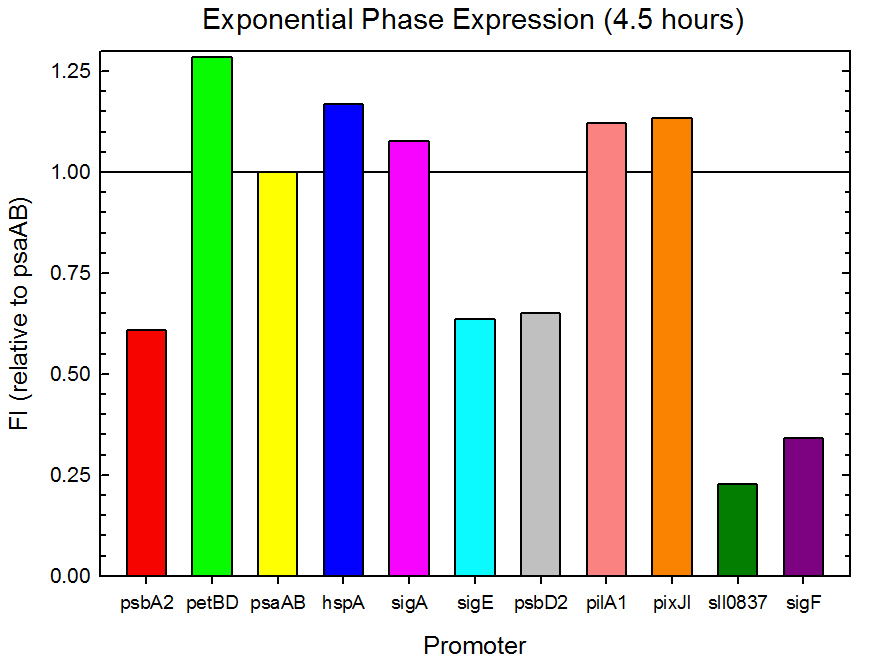
| - | Figure 2. Exponential phase GFP expression relative to psaAB expression for 11 ''Synechocystis'' promoters. Measurements were from the middle of exponential phase, 4.5 hours into the experiment. | + | '''Figure 2.''' Exponential phase GFP expression relative to psaAB expression for 11 ''Synechocystis'' promoters. Measurements were from the middle of exponential phase, 4.5 hours into the experiment. |
[[Image:USU_Stationary_Phase.JPG||600px]] | [[Image:USU_Stationary_Phase.JPG||600px]] | ||
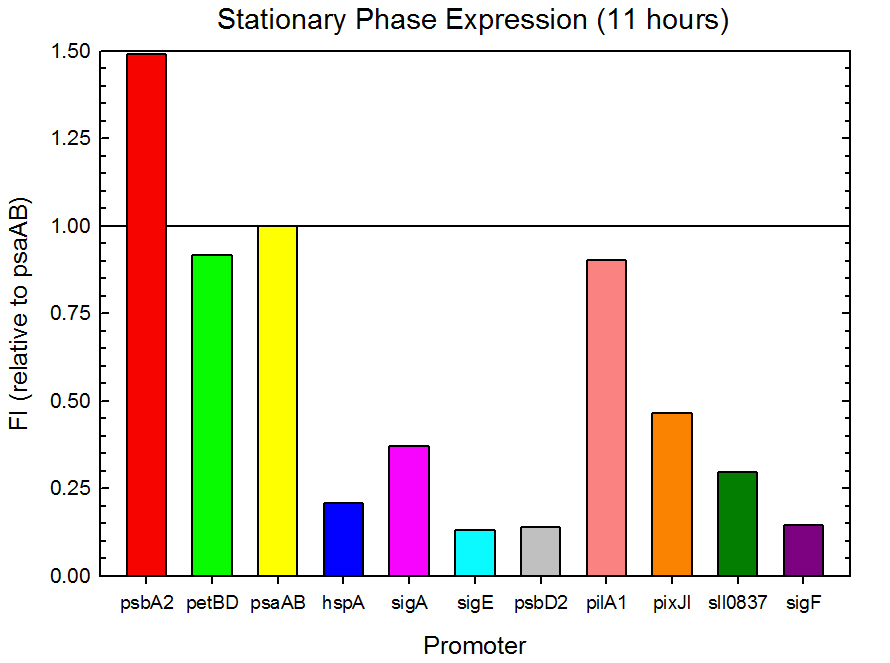
| - | Figure 3. Stationary phase GFP expression relative to psaAB expression for 11 ''Synechocystis'' promoters. Measurements were from the middle of the stationary phase, 11 hours into the experiment. | + | '''Figure 3.''' Stationary phase GFP expression relative to psaAB expression for 11 ''Synechocystis'' promoters. Measurements were from the middle of the stationary phase, 11 hours into the experiment. |
==Expression Results== | ==Expression Results== | ||
| - | As expected from the high homology from promoter and RBS alignments (Tables | + | As expected from the high homology from promoter and RBS alignments (Tables 2 and 3, above), the Type I promoters possessed the highest level of GFP in both growth phases. Interestingly, expression levels of ''hsp''A and ''sig''A were high during exponential phase and decreased significantly during stationary phase. Type II and Type III promoters exhibited moderate to low expression in both phases, likely due to the reduced homology with the consensus sequences. Despite low homology with the sigma70 promoter sequences, ''pil''A1 exhibited high expression in both growth phases, and ''pix''JI exhibited high expression in exponential phase. This could potentially be due to homology to other sigma factor binding sequences in ''E. coli'', but these alignments have not yet been explored. The final expression levels are listed in Table 4 below for easier reference. |
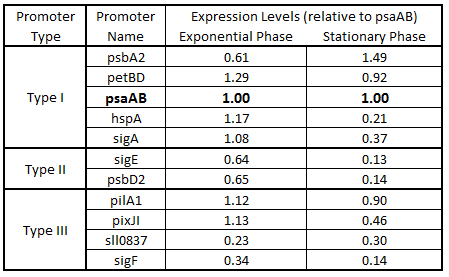
| - | Table | + | '''Table 4.''' Expression levels of 11 ''Synechocystis'' promoters in ''E. coli'' during exponential and stationary phase. Values are shown relative to ''psa''AB expression levels. |
[[Image:USU_Promoter_Expression.png]] | [[Image:USU_Promoter_Expression.png]] | ||
| Line 60: | Line 63: | ||
=Team Accomplishments= | =Team Accomplishments= | ||
==Bronze Medal== | ==Bronze Medal== | ||
| - | 70 BioBrick Parts were designed including a platform for introducing BioBrick constructs into species that accept DNA through homologous recombination. Project design resulted from a literature review performed by student participants, building primarily on the work of Kunert (2000) and Asayama ( | + | <uL> |
| + | <li>70 BioBrick Parts were designed including a platform for introducing BioBrick constructs into species that accept DNA through homologous recombination. Project design resulted from a literature review performed by student participants, building primarily on the work of Kunert, et al. (2000) and Immamura and Asayama (2009). All work was performed by student participants outside of primer oligo synthesis and DNA sequencing. Safety considerations are addressed in our Lab section. | ||
| - | 43 constructed BioBrick Parts were submitted to the parts registry. | + | <li>43 constructed BioBrick Parts were submitted to the parts registry. |
| - | Plasmid pSB1A3 was mutated to include restriction sites creating a platform for efficient genome modification by homologous recombination in species outside of ''E. coli''. Structure was verified by restriction enzyme digestion. | + | <li>Plasmid pSB1A3 was mutated to include restriction sites creating a platform for efficient genome modification by homologous recombination in species outside of ''E. coli''. Structure was verified by restriction enzyme digestion. |
| - | 15 promoters with 5'UTR and RBS were isolated from the genome of ''Synechocystis'' PCC 6803, converted to BioBrick format, and submitted in pSB1C3. Parts composed of the promoters without the 5'UTR and RBS were constructed as well as a 5'UTR and RBS cloned upstream of GFP which will allow us to test scar effects in ''Synechocystis'' in the future. | + | <li>15 promoters with 5'UTR and RBS were isolated from the genome of ''Synechocystis'' PCC 6803, converted to BioBrick format, and submitted in pSB1C3. Parts composed of the promoters without the 5'UTR and RBS were constructed as well as a 5'UTR and RBS cloned upstream of GFP which will allow us to test scar effects in ''Synechocystis'' in the future. |
| + | </uL> | ||
==Silver medal== | ==Silver medal== | ||
| - | While functionality of these parts in ''Synechocystis'' has not yet been characterized, expression of these parts cloned upstream of GFP was analyzed in ''E. coli'' as described above and functioned as expected. | + | <uL> |
| + | <li>While functionality of these parts in ''Synechocystis'' has not yet been characterized, expression of these parts cloned upstream of GFP was analyzed in ''E. coli'' as described above and functioned as expected. | ||
| - | Part BBa_K390501 functions as described in our Construction page. | + | <li>Part BBa_K390501 functions as described in our Construction page, effectively secreting a target protein complexed with bioplastic granules into the extracellular media. 1H NMR was used as the diagnostic tool. |
| + | </uL> | ||
==Gold Medal== | ==Gold Medal== | ||
| - | Part BBa_K208029 created by the USU 2009 iGEM team was further characterized and functionality confirmed and improved by creating a new composite part as described in the Construction page. | + | <uL> |
| + | <li>Part BBa_K208029 created by the USU 2009 iGEM team was further characterized and functionality confirmed and improved by creating a new composite part as described in the Construction page. | ||
| - | Our 2010 USU iGEM team collaborated with the iGEM Osaka 2010 team through the contact Teo Shao Thing and provided BioBrick parts requested by team Osaka. | + | <li>Our 2010 USU iGEM team collaborated with the iGEM Osaka 2010 team through the contact Teo Shao Thing and provided BioBrick parts requested by team Osaka. |
| - | Bioinformatics software was developed by our team to analyze upstream regions of genes. This provided information about ''Synechocystis'' PCC 6803 that was not available using commercially available software. This software could be used to analyze other genomes and also can be used for codon optimization in gene synthesis. | + | <li>Bioinformatics software was developed by our team to analyze upstream regions of genes. This provided information about ''Synechocystis'' PCC 6803 that was not available using commercially available software. This software could be used to analyze other genomes and also can be used for codon optimization in gene synthesis. |
| + | </uL> | ||
</div> | </div> | ||
{{:Team:Utah_State/usu_footer}} | {{:Team:Utah_State/usu_footer}} | ||
Latest revision as of 00:44, 28 October 2010
Results
Contents |
Synechocystis Promoter Expression in E. coli
Promoter regions, 5’ Untranslated regions (5’UTR), and ribosome binding sites (RBS) from 11 Synechocystis genes were cloned upstream of GFP (Table 1). While these parts are eventually going to be characterized and used in Synechocystis, since the final integration plasmid was not complete, we decided to test these parts in E. coli. We believed that these parts would function in E. coli due to the similarity of the sigma factor binding sites and RBS sequences from Synechocystis to the consensus promoter and RBS in E. coli (Tables 2 and 3).
Table 1.Table of BioBrick Part Numbers and Promoters.
Table 2. Sigma factor binding site alignments. Underlined sequences are the binding region for sigma70 sigma factor from E. coli. Bases in red are exact matches to the consensus sequence.
Table 3. Ribosome binding site sequence alignments. The regions 25 bp upstream of the start codon for each gene were aligned to the E. coli consensus RBS sequence. Underlined sequences are the binding region for the 9 bp 3’ end of the 16S rRNA subunit in E. coli. Bases in red are exact matches to the consensus sequence.
Preliminary Analysis
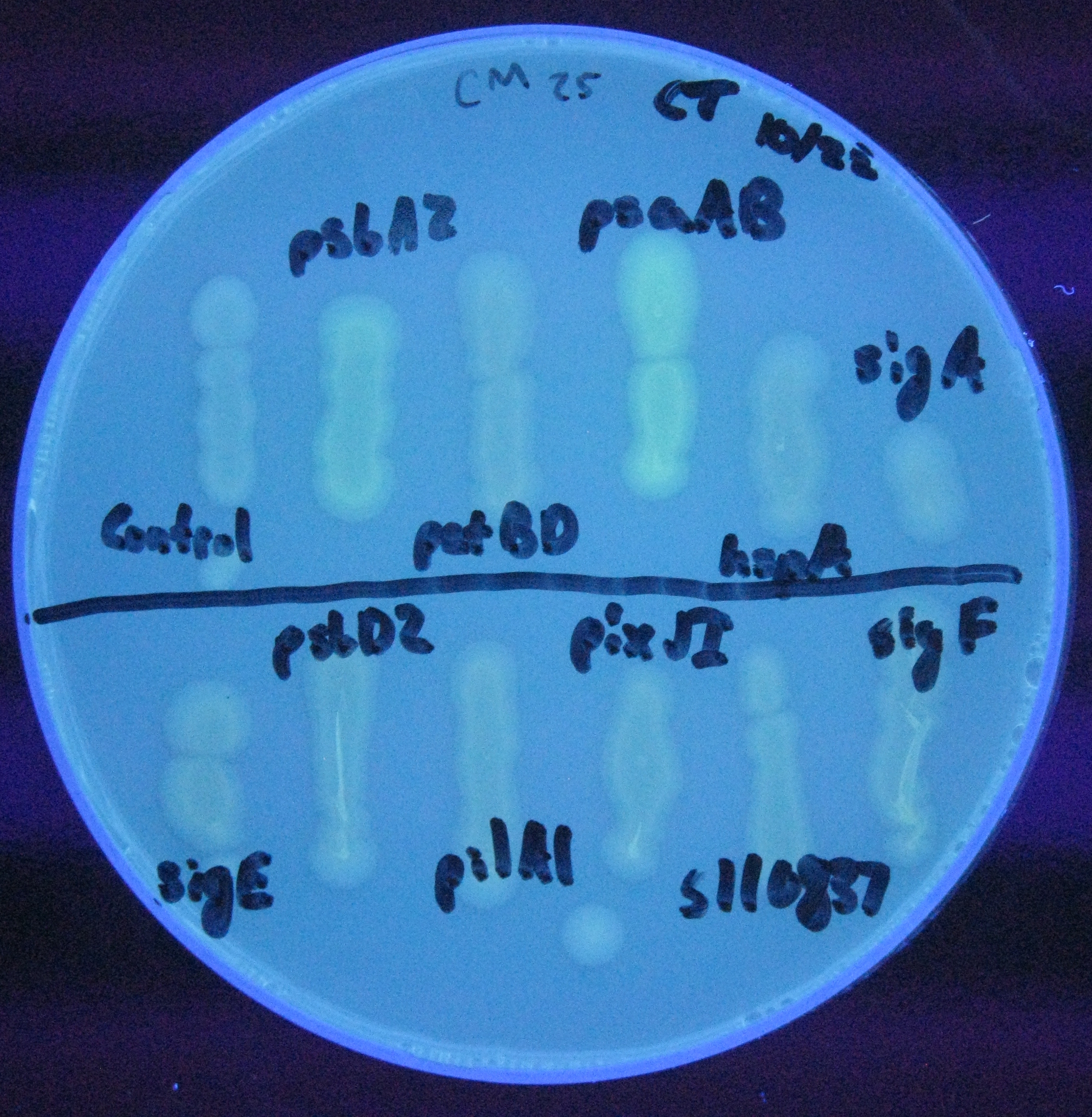
For our initial analysis, we plated small amounts of each of the transformed cells onto LB agar plates, and visually checked for GFP expression after 24 hours growth. GFP expression was clearly observed in the psaAB and psbA2 constructs, with the psaAB construct exhibiting the highest level of fluorescence (Figure 1).
Figure 1. Cell plating of Synechocystis promoters driving GFP expression in E. coli.
Fluorometer Analysis
Fluorometer analysis of the 11 Synechocystis promoter, 5’UTR, and RBS regions in front of GFP was conducted as described in the Procedures section. Measurements were taken every 30 minutes and expression levels at the midpoints of exponential and stationary phase are shown in Figures 2 and 3. Since psaAB exhibited fluorescence during the preliminary analysis and was observed to be consistent with time across the fluorometer measurements, it was used as the standard, and all fluorescence values were calculated relative to its expression level.
Figure 2. Exponential phase GFP expression relative to psaAB expression for 11 Synechocystis promoters. Measurements were from the middle of exponential phase, 4.5 hours into the experiment.
Figure 3. Stationary phase GFP expression relative to psaAB expression for 11 Synechocystis promoters. Measurements were from the middle of the stationary phase, 11 hours into the experiment.
Expression Results
As expected from the high homology from promoter and RBS alignments (Tables 2 and 3, above), the Type I promoters possessed the highest level of GFP in both growth phases. Interestingly, expression levels of hspA and sigA were high during exponential phase and decreased significantly during stationary phase. Type II and Type III promoters exhibited moderate to low expression in both phases, likely due to the reduced homology with the consensus sequences. Despite low homology with the sigma70 promoter sequences, pilA1 exhibited high expression in both growth phases, and pixJI exhibited high expression in exponential phase. This could potentially be due to homology to other sigma factor binding sequences in E. coli, but these alignments have not yet been explored. The final expression levels are listed in Table 4 below for easier reference.
Table 4. Expression levels of 11 Synechocystis promoters in E. coli during exponential and stationary phase. Values are shown relative to psaAB expression levels.
Team Parts
<groupparts>iGEM010 Utah_State</groupparts>
Team Accomplishments
Bronze Medal
- 70 BioBrick Parts were designed including a platform for introducing BioBrick constructs into species that accept DNA through homologous recombination. Project design resulted from a literature review performed by student participants, building primarily on the work of Kunert, et al. (2000) and Immamura and Asayama (2009). All work was performed by student participants outside of primer oligo synthesis and DNA sequencing. Safety considerations are addressed in our Lab section.
- 43 constructed BioBrick Parts were submitted to the parts registry.
- Plasmid pSB1A3 was mutated to include restriction sites creating a platform for efficient genome modification by homologous recombination in species outside of E. coli. Structure was verified by restriction enzyme digestion.
- 15 promoters with 5'UTR and RBS were isolated from the genome of Synechocystis PCC 6803, converted to BioBrick format, and submitted in pSB1C3. Parts composed of the promoters without the 5'UTR and RBS were constructed as well as a 5'UTR and RBS cloned upstream of GFP which will allow us to test scar effects in Synechocystis in the future.
Silver medal
- While functionality of these parts in Synechocystis has not yet been characterized, expression of these parts cloned upstream of GFP was analyzed in E. coli as described above and functioned as expected.
- Part BBa_K390501 functions as described in our Construction page, effectively secreting a target protein complexed with bioplastic granules into the extracellular media. 1H NMR was used as the diagnostic tool.
Gold Medal
- Part BBa_K208029 created by the USU 2009 iGEM team was further characterized and functionality confirmed and improved by creating a new composite part as described in the Construction page.
- Our 2010 USU iGEM team collaborated with the iGEM Osaka 2010 team through the contact Teo Shao Thing and provided BioBrick parts requested by team Osaka.
- Bioinformatics software was developed by our team to analyze upstream regions of genes. This provided information about Synechocystis PCC 6803 that was not available using commercially available software. This software could be used to analyze other genomes and also can be used for codon optimization in gene synthesis.
 "
"