Team:Slovenia/PROJECT/proof/popfret/split
From 2010.igem.org
| (32 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<style> | <style> | ||
#vsebina_mid{ | #vsebina_mid{ | ||
| - | height: | + | height: 3500px; |
} | } | ||
#lgumb2{ | #lgumb2{ | ||
background-image:url("https://static.igem.org/mediawiki/2010/3/3e/SLOproofover.png"); | background-image:url("https://static.igem.org/mediawiki/2010/3/3e/SLOproofover.png"); | ||
| + | } | ||
| + | #subgumb2b{ | ||
| + | |||
| + | background-image:url("https://static.igem.org/mediawiki/2010/6/67/SLOpopfretover.png"); | ||
} | } | ||
#subgumb3{display:none}#subgumb4{display:none}#subgumb5{display:none}#subgumb6{display:none}#subgumb7{display:none}#subgumb8{display:none}#subgumb9{display:none}#subgumb10{display:none} | #subgumb3{display:none}#subgumb4{display:none}#subgumb5{display:none}#subgumb6{display:none}#subgumb7{display:none}#subgumb8{display:none}#subgumb9{display:none}#subgumb10{display:none} | ||
| Line 51: | Line 55: | ||
<div id="overhead"> | <div id="overhead"> | ||
| + | |||
| + | |||
| + | <span style="width:322px;" id="naslov">split GFP reconstitution </span> | ||
| + | </div> | ||
| + | <div id="thumbsi"> | ||
<a href="/Team:Slovenia/PROJECT/proof/popfret/back"><span style="font-size:15px; | <a href="/Team:Slovenia/PROJECT/proof/popfret/back"><span style="font-size:15px; | ||
| - | width:200px;" id="stopnja3"> | + | width:200px;" id="stopnja3">background</span></a> |
<a href="/Team:Slovenia/PROJECT/proof/popfret/split"><span style="font-size:15px; | <a href="/Team:Slovenia/PROJECT/proof/popfret/split"><span style="font-size:15px; | ||
| - | width:200px;" id="stopnja3a"> | + | width:200px;" id="stopnja3a">split GFP reconstitution</span></a> |
| - | <a href="/Team:Slovenia/PROJECT/proof/popfret/fret"><span style="font-size:15px; | + | <a href="/Team:Slovenia/PROJECT/proof/popfret/fret"><span style="font-size:15px;width:200px;" id="stopnja3">quadruple binding</span></a> |
| - | width:200px;" id="stopnja3"> | + | |
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div id="besedilo"> | <div id="besedilo"> | ||
| Line 67: | Line 73: | ||
__TOC__ | __TOC__ | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
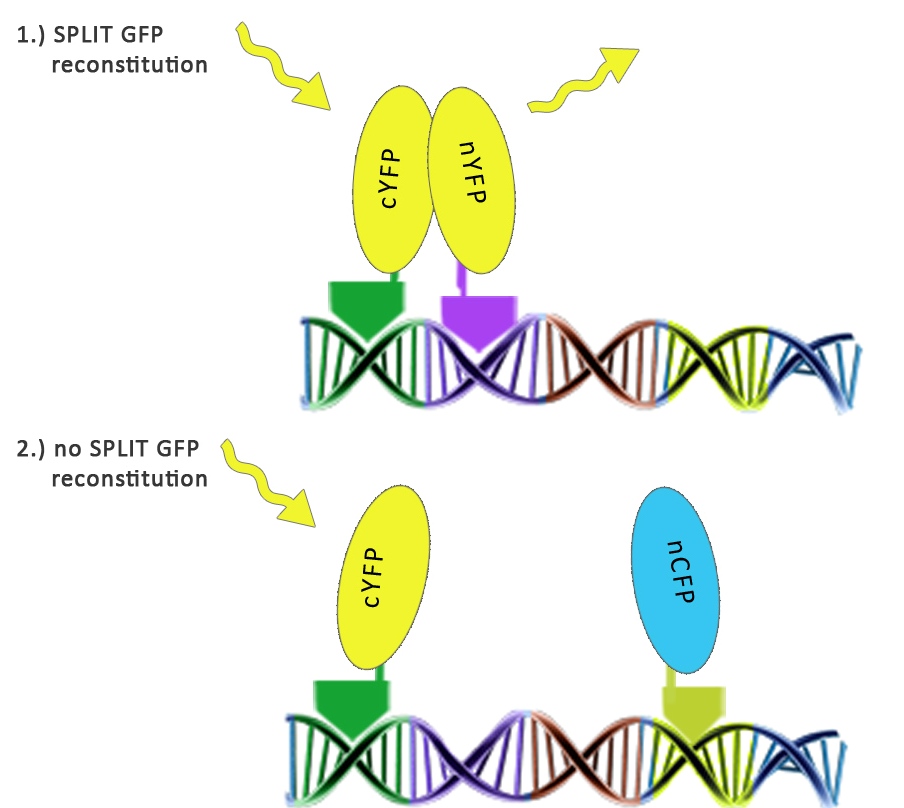
| - | We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated <em>in vivo</em>, but only <em>in vitro </em>(Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly<em> in vitro</em> using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus. | + | We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated <em>in vivo</em>, but only <em>in vitro </em>(Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly<em> in vitro</em> using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus.<br><br> |
| - | [[Image:slo_split_prava.jpg|thumbs|center| | + | [[Image:slo_split_prava.jpg|thumbs|center|500px]] |
| + | <br><br> | ||
<h2>Results</h2> | <h2>Results</h2> | ||
| - | |||
| - | |||
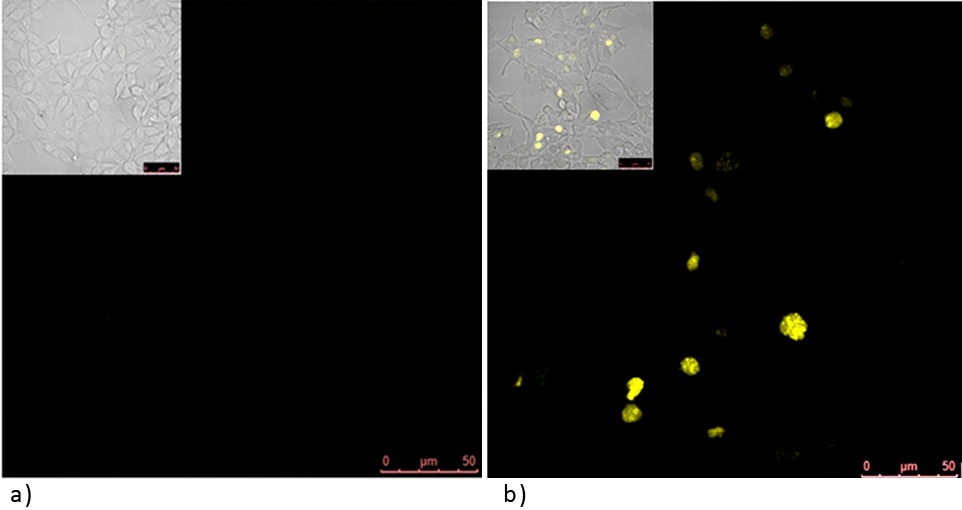
| - | Split YFP reconstitution occurs only in the presence of the appropriate DNA program. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268, 25ng each, were cotransfected into HEK293 cells in the (a) absence and (b) presence of 500 ng of DNA program sequence in [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 pSB1C3 backbone]. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm. | + | Experiments below show how a DNA program sequence is involved in reconstitution of a split fluorescent protein, in our case being mCitrine due to its enhanced stability over other YFP variants. What we have shown is: DNA program mediates spacer length dependent reconstitution of a split fluorescent protein into its mature functional form. |
| + | |||
| + | [[Image:Slo split test.png|thumbs|center|550px]] | ||
| + | |||
| + | '''Figure 1: Split YFP reconstitution occurs only in the presence of the appropriate DNA program. '''[http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268], 25ng each, were cotransfected into HEK293 cells in the (a) absence and (b) presence of 500 ng of DNA program sequence in [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 pSB1C3 backbone]. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm.<br><br> | ||
| + | |||
| + | [[Image:Slo split rast.png|thumbs|center|700px]] | ||
| + | |||
| + | |||
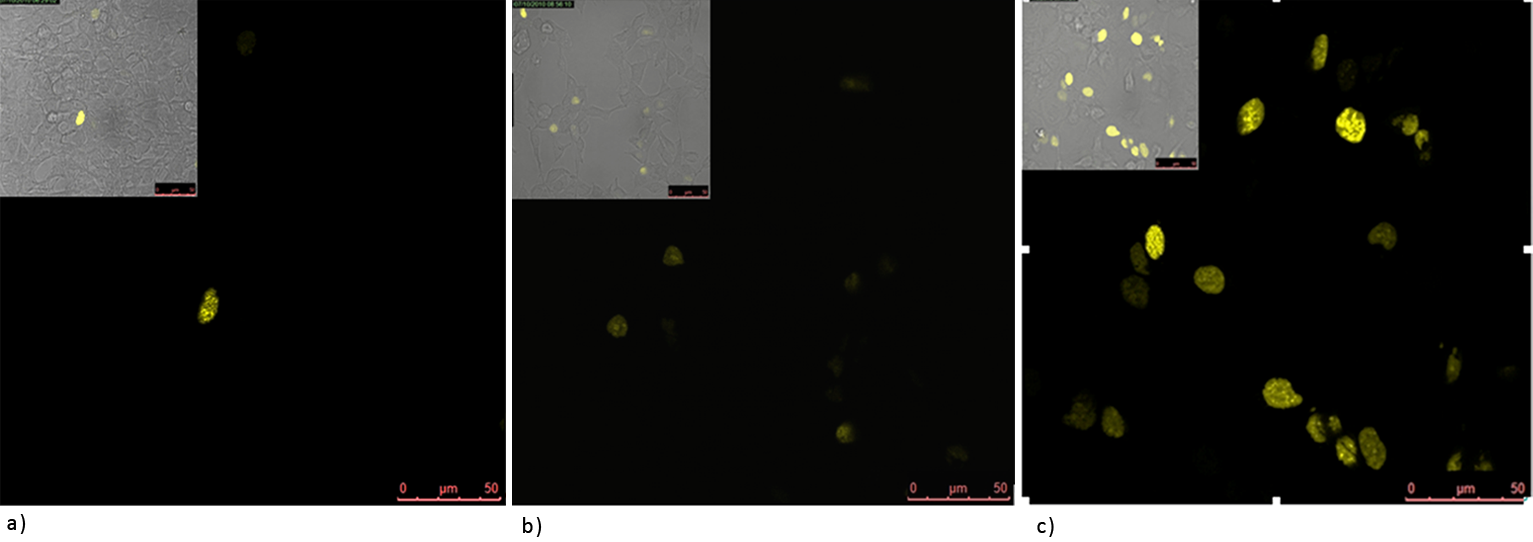
| + | '''Figure 2: Split YFP reconstitution efficiency depends on the amount of a DNA program.''' HEK293 cells were co-transfected with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268] (50ng each) and different amount of DNA program [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039]) (a) 50ng (b) 100ng and (c) 200ng. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm.<br><br> | ||
| + | |||
| + | [[Image:test_razdalje.png|thumbs|center|400px]] | ||
| + | <br> | ||
| + | |||
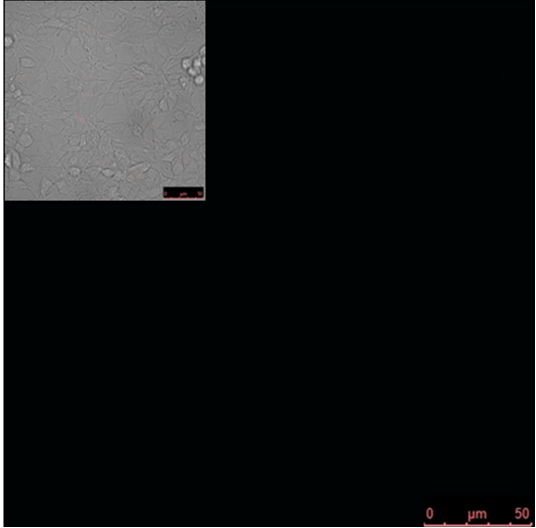
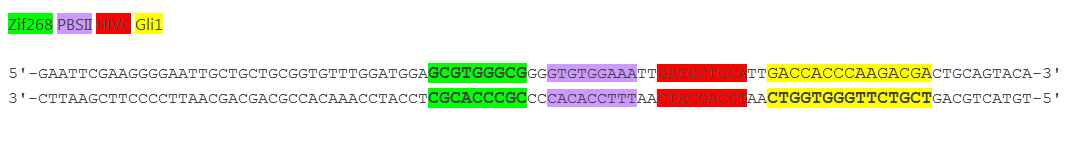
| + | '''Figure 3: Increased distance between binding sites for zinc finger split fluorescent protein fusions prevents the reassembly of fluorescent proteins.''' HEK293 cells were cotransfected with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323016 Gli1_link_nCFP] and 280ng of DNA program [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039]. Binding elements for DNA binding factors of chimeric proteins, split GFPs, were separated by 2 other zinc finger operators - HIVC_O and PBSII_O (shematic presentation above). The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm. The fluorescence was not detected, demonstrating that the split GFP reassembly depends on the distance between zinc finger binding sites on the DNA program sequence. Binding sites of fusion proteins are indicated below [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039]. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:SLO_fretkode2.png|thumbs|center|650px]] | ||
| + | <br><br> | ||
| + | [[Image:SLOinvitro.png|thumbs|center|600px]] | ||
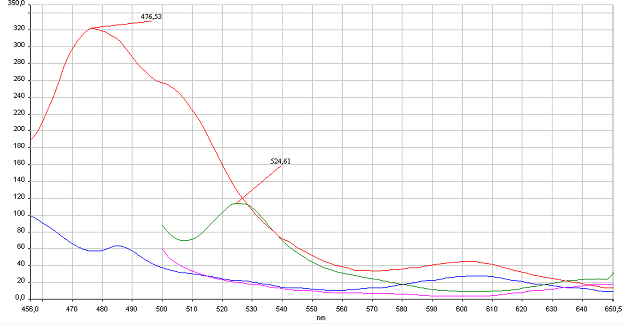
| + | '''Figure 4: Reconstitution of split CFP and YFP <em>in vitro</em> is based on binding to DNA program.''' HEK293 were cotransfected with 1 µg of plasmids of split pairs; [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323021 Gli1_link_nCFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323029 cCFP_link_HIVC], and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268]. Cells were lysed using Promega Passive Lysis Buffer. Lysates carrying split GFP halves were incubated in the presence of DNA program sequence, which was generated with annealing of complementary p</span>rimers, at 4°C overnight. Spectra were acquired using Perkin Elmer Instruments LS55 Luminescence Spectrometer. Blue and violet line: lysate of untransfected cells excited at 433 nm and 480 nm, respectively. Red and green curves represent spectra of reconstituted mCerulean and mCitrine respectively. It can be seen from emission peaks of lysate spectra that the split GFPs form mature GFPs in the presence of program DNA also <em>in vitro</em>.<br> | ||
| + | <h2>Discussion</h2> | ||
| + | Experiments confirmed our initial hypothesis and showed that DNA program sequence can be used as a scaffold assembling chimeric proteins consisting of DNA binding domain and functional domains e.g. split GFP. Undoubtedly, the reconstitution of split GFPs linked to zink fingers was regulated by DNA program sequence. We also observed that the number of cells having reassembled GFP <em>in vivo</em> depends on the amount of DNA program added to the cells. Experiments underlie the idea that spacer distance between DNA binding elements is important in mediating GFP reconstitution. | ||
<!--STOP BESEDILO--> | <!--STOP BESEDILO--> | ||
Latest revision as of 22:22, 27 October 2010
Contents |
Introduction
We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated in vivo, but only in vitro (Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly in vitro using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus.
Results
Experiments below show how a DNA program sequence is involved in reconstitution of a split fluorescent protein, in our case being mCitrine due to its enhanced stability over other YFP variants. What we have shown is: DNA program mediates spacer length dependent reconstitution of a split fluorescent protein into its mature functional form.
Figure 1: Split YFP reconstitution occurs only in the presence of the appropriate DNA program. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268], 25ng each, were cotransfected into HEK293 cells in the (a) absence and (b) presence of 500 ng of DNA program sequence in [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 pSB1C3 backbone]. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm.
Figure 2: Split YFP reconstitution efficiency depends on the amount of a DNA program. HEK293 cells were co-transfected with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268] (50ng each) and different amount of DNA program [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039]) (a) 50ng (b) 100ng and (c) 200ng. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm.
Figure 3: Increased distance between binding sites for zinc finger split fluorescent protein fusions prevents the reassembly of fluorescent proteins. HEK293 cells were cotransfected with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323016 Gli1_link_nCFP] and 280ng of DNA program [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039]. Binding elements for DNA binding factors of chimeric proteins, split GFPs, were separated by 2 other zinc finger operators - HIVC_O and PBSII_O (shematic presentation above). The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm. The fluorescence was not detected, demonstrating that the split GFP reassembly depends on the distance between zinc finger binding sites on the DNA program sequence. Binding sites of fusion proteins are indicated below [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 BBa_323039].
Figure 4: Reconstitution of split CFP and YFP in vitro is based on binding to DNA program. HEK293 were cotransfected with 1 µg of plasmids of split pairs; [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323021 Gli1_link_nCFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323029 cCFP_link_HIVC], and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268]. Cells were lysed using Promega Passive Lysis Buffer. Lysates carrying split GFP halves were incubated in the presence of DNA program sequence, which was generated with annealing of complementary p</span>rimers, at 4°C overnight. Spectra were acquired using Perkin Elmer Instruments LS55 Luminescence Spectrometer. Blue and violet line: lysate of untransfected cells excited at 433 nm and 480 nm, respectively. Red and green curves represent spectra of reconstituted mCerulean and mCitrine respectively. It can be seen from emission peaks of lysate spectra that the split GFPs form mature GFPs in the presence of program DNA also in vitro.
Discussion
Experiments confirmed our initial hypothesis and showed that DNA program sequence can be used as a scaffold assembling chimeric proteins consisting of DNA binding domain and functional domains e.g. split GFP. Undoubtedly, the reconstitution of split GFPs linked to zink fingers was regulated by DNA program sequence. We also observed that the number of cells having reassembled GFP in vivo depends on the amount of DNA program added to the cells. Experiments underlie the idea that spacer distance between DNA binding elements is important in mediating GFP reconstitution.
 "
"