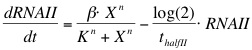Team:Groningen/Killswitch model
From 2010.igem.org
(→The model) |
(→Kill Switch) |
||
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
==Kill Switch== | ==Kill Switch== | ||
| + | <html> | ||
| + | <div style="text-align: justify"> | ||
| + | </html> | ||
For safety reasons a kill switch is needed in our project. When our hydrofobic biofilm would be applied as a coating, the cells should be programmed to die after the expression of the chaplins. In safety regulations only the release of living genetically modified organisms is a problem. So with a kill switch, the applications for our biofilm will be broader and easier to realize. Different types of kill switches are found in nature. Some of these switches are briefly discussed below. | For safety reasons a kill switch is needed in our project. When our hydrofobic biofilm would be applied as a coating, the cells should be programmed to die after the expression of the chaplins. In safety regulations only the release of living genetically modified organisms is a problem. So with a kill switch, the applications for our biofilm will be broader and easier to realize. Different types of kill switches are found in nature. Some of these switches are briefly discussed below. | ||
| + | |||
===Toxin/antidote system=== | ===Toxin/antidote system=== | ||
| - | One | + | <html> |
| - | different addiction systems are found for different low copy plasmids. All those systems are based on a stable toxin and a proteolytically unstable antidote. | + | <div style="text-align: justify"> |
| + | </html> | ||
| + | One possibility would be to modify the plasmid addiction system of bacteriophage P1 which consists of a toxin (Doc) and an antidote (Phd). Cells that have lost the P1 genome will die because the antidote is gradually degraded while the toxin is stable. Gazit et. al. suggests two different functions of the antidote. First, it is possible that it binds the toxin and physically blocks the interaction of the toxin with the target molecules or by altering the structure of the toxin and thereby the toxin activity. The second option is that it activates another protein that neutralizes the toxin. In ''E. coli'' different addiction systems are found for different low copy plasmids. All those systems are based on a stable toxin and a proteolytically unstable antidote. In order to use this mechanism as a kill switch, the production of antidote would have to be suppressed during late stage stationary phase which would lead to the death off these cells after the formation off the hydrophobic biofilm. | ||
| - | |||
| - | |||
| - | |||
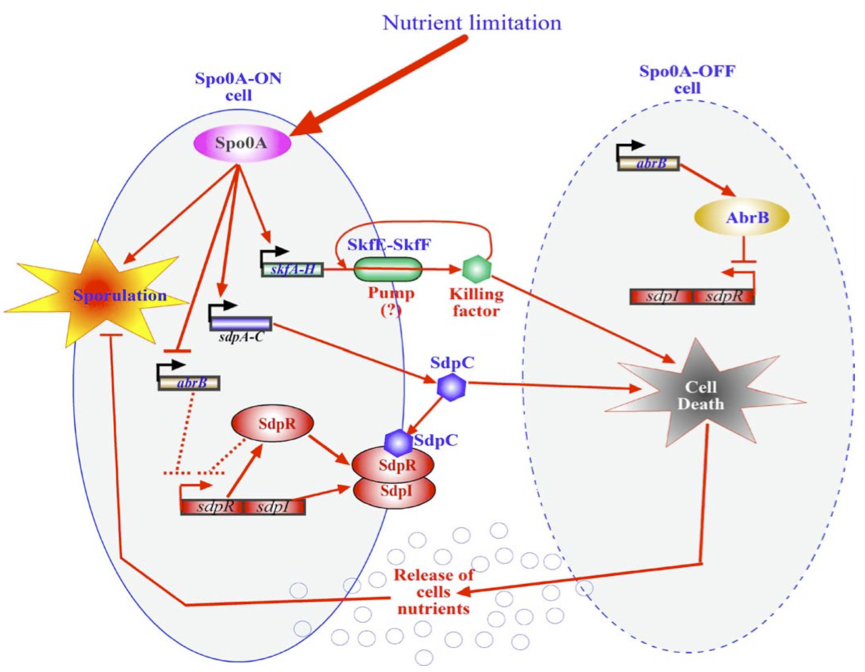
| - | [[Image:Groningen-Kulka.png|500px|Schematic representation of the killing mechanism in Bacillus subtilis]] | + | ===Killswitch coupled to sporulation in ''Bacillus subtilis''=== |
| + | <html> | ||
| + | <div style="text-align: justify"> | ||
| + | </html> | ||
| + | [[Image:Groningen-Kulka.png|500px|Schematic representation of the killing mechanism in Bacillus subtilis|right]] | ||
| + | Another way of killing the cells once the biofilm has formed would be to alter an already existing in ''B. subtillis'' Within ''Bacillus subtilis'' a system exists whereby the sporulation process is delayed by killing surrounding cells. Cells that are Spo0A-Off are lysed whereby nutrients are released. These nutrients can be consumed by the Spo0A-On | ||
| + | cells thereby delaying the process of sporulation The skf and sdp operons are responsible for the death of a subpopulation of sporulating bacterial cells. The regulatory protein Spo0A up regulates the sporulating killing factor (skfA-H operon) and the sporulating delay protein (sdpABC operon). Surrounding cells that are Spo0A-Off are killed by the extracellular killing factor skf, produced by Spo0A-On cells. SkfE and SkfF together probably act as an export pump that secretes the killing factor and thereby protect the cell. Another killing factor is sdpC. The Spo0A-On cells have two mechanisms that provide resistance to sdpC. The first one is the SdpC-SdpI-SdpR signaling pathway. The second is the repression of AbrB synthesis. A schematic overview of this mechanism is given below. If any of the factors that protect the cells from their own toxins were dysfunctional, the cells would kill themselves as soon as the Spo0A system would be turned on due nutrient limitation (which will happen inevitably in a biofilm after a while). | ||
| + | |||
===Anti-sense RNA regulation=== | ===Anti-sense RNA regulation=== | ||
| Line 18: | Line 28: | ||
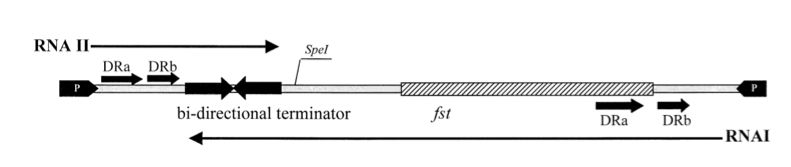
[[Image:Groningen-Greenfield2000.png|500px|Features of the pAD1 par stability determinant.]] | [[Image:Groningen-Greenfield2000.png|500px|Features of the pAD1 par stability determinant.]] | ||
| + | <html> | ||
| + | <div style="text-align: justify"> | ||
| + | </html> | ||
Promoters for RNA I and RNA II are indicated by black arrows on each end. The open reading frame, fst, encodes for a 33-amino-acid peptide. The two RNAs read in opposite directions across direct repeats labelled DRa and DRb (arrows) to a bidirectional terminator (converging arrows) (Greenfield, 2000). | Promoters for RNA I and RNA II are indicated by black arrows on each end. The open reading frame, fst, encodes for a 33-amino-acid peptide. The two RNAs read in opposite directions across direct repeats labelled DRa and DRb (arrows) to a bidirectional terminator (converging arrows) (Greenfield, 2000). | ||
The so-called par region encodes for two small RNAs , RNAI and RNAII as depicted in the above figure. RNAI is 210nt long and contains the fst gene which encodes the toxic compound. RNAII is only 65nt long and highly complementary to RNAI. | The so-called par region encodes for two small RNAs , RNAI and RNAII as depicted in the above figure. RNAI is 210nt long and contains the fst gene which encodes the toxic compound. RNAII is only 65nt long and highly complementary to RNAI. | ||
| - | This results in complex formation whereby RNAI is not able to translate the fst gene into the faecalis plasmid-stabilizing toxin compound. The half-life time of RNAI is only 10 minutes while the half-life time of RNAII is more than one hour. (Zielenkiewicz)(Greenfield) This difference can be used as some sort of timer on a killswitch. | + | This results in complex formation whereby RNAI is not able to translate the fst gene into the faecalis plasmid-stabilizing toxin compound. The half-life time of RNAI is only 10 minutes while the half-life time of RNAII is more than one hour. (Zielenkiewicz)(Greenfield) This difference can be used as some sort of timer on a killswitch since the toxin concentration will eventually outcompete the antitoxin concentration. Our model is based on this kill switch system. |
| - | Our model is based on this kill switch system. | + | |
===Promoters=== | ===Promoters=== | ||
| - | + | <html> | |
| - | promoter | + | <div style="text-align: justify"> |
| + | </html> | ||
| + | Regulating the kill switch with the same promoters used for chaplin expression would ensure that the cells die after the job is done (the unstable antitoxin takes some time to loose its effect). Hence the srfA and yqxM promoters which are also [https://2010.igem.org/Team:Groningen#/biofilm considered] for chaplin expression would be used to initiate the kill switch too. The srfA promoter is activated already in the early stages of biofilm formation whereas the yqxM promoter is only activated from the beginning of the stationary phase. | ||
| + | |||
| + | To ensure a bigger time delay between chaplin expression and kill switch activation, the srfA promoter could start the chaplin expression, whereas the yqxM promoter which is active later in the growth curve would start the kill switch. | ||
| - | |||
The activation of the par region can be modeled with a simple activator term. The degradation of the two RNAs can be modelled with half life equations. The change in biomass can be modeled with a simple growth model combined with | The activation of the par region can be modeled with a simple activator term. The degradation of the two RNAs can be modelled with half life equations. The change in biomass can be modeled with a simple growth model combined with | ||
the delta tox. | the delta tox. | ||
| + | |||
===The model=== | ===The model=== | ||
The formation of RNAI and RNAII is described with the Hill's equation. The degradation of the RNA is taken into account by using the half-life of the different RNA's. The Hill's equation is the same for both RNA's because the assumption is made that both promoters are equal in strength. | The formation of RNAI and RNAII is described with the Hill's equation. The degradation of the RNA is taken into account by using the half-life of the different RNA's. The Hill's equation is the same for both RNA's because the assumption is made that both promoters are equal in strength. | ||
| - | [[Image:Eq_RNAI_Killswitch. | + | [[Image:RUG Eq_RNAI_Killswitch.jpg]] |
| - | [[Image:Eq_RNAII_Killswitch. | + | [[Image:RUG Eq_RNAII_Killswitch.jpg]] |
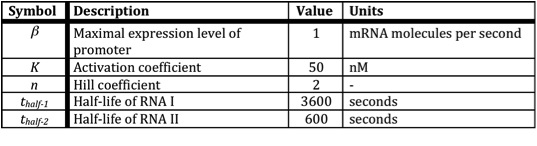
| - | + | In the following table all constants used in the model are given. | |
| - | The assumption is made that the promoter is very strong, therefore the maximal expression level of the promoter is assumed to be 1 mRNA molecule per second. Normally promoter expression levels range between | + | [[Image:RUG Table_Killswitch.jpg]] |
| + | |||
| + | The assumption is made that the promoter is very strong, therefore the maximal expression level of the promoter is assumed to be 1 mRNA molecule per second. Normally promoter expression levels range between 10-4 and 1 mRNA molecules per second. | ||
K is the activation coefficient and defines the concentration active X needed to significantly activate expression. The concentration of transcription factor is assumed to be much higher than the activation coefficient K. A value of 500 nM is chosen for this model. | K is the activation coefficient and defines the concentration active X needed to significantly activate expression. The concentration of transcription factor is assumed to be much higher than the activation coefficient K. A value of 500 nM is chosen for this model. | ||
| - | + | [[Image: RUG killswitch_Laura.jpg]] | |
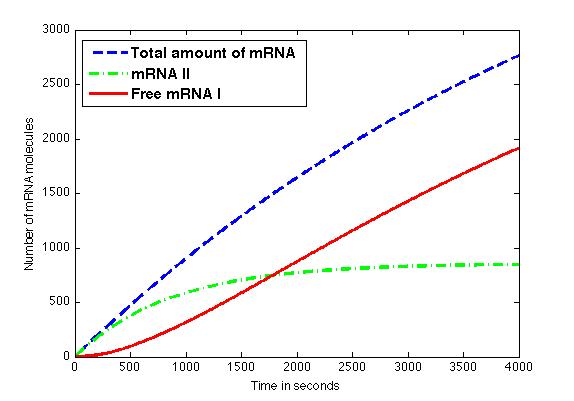
The figure shows the amount of mRNA molecules in time. Assuming that all available mRNA II binds to mRNA I, the red line in the graph shows the amount of free mRNA I molecules. Translation of these mRNA I molecules results in the formation of a toxic compound. Above a certain threshold this will result in cell death. | The figure shows the amount of mRNA molecules in time. Assuming that all available mRNA II binds to mRNA I, the red line in the graph shows the amount of free mRNA I molecules. Translation of these mRNA I molecules results in the formation of a toxic compound. Above a certain threshold this will result in cell death. | ||
| + | |||
| + | |||
| + | ===Matlab source files=== | ||
| + | Killswitch ([http://dl.dropbox.com/u/391356/iGEM/killswitchMatlab.zip source]) | ||
| + | |||
===References=== | ===References=== | ||
| - | <small> | + | <small>Gazit E and Sauer RT, The Doc Toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. Journal of Biological Chemistry, 1999, 274 (24):16813-8. PMID 10358024 |
| - | + | ||
| - | Greenfield | + | Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE, The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism, Molecular Microbiology, 2000, 37(3): 652-660. PMID 10931358 |
| - | + | Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R, Bacterial programmed cell death and multicellular behavior in bacteria, Plos genetics 2(10): e135. PMID 15036322 | |
| - | + | Zielenkiewicz U and Ceglowski P, Mechanisms of plasmid stable maintenance with special focus on plasmid addiction ystems, Acta Biochimica Polonia, 2001, 48 (4): 1003-1023. PMID 11995964</small> | |
| - | </small> | + | |
Latest revision as of 19:25, 27 October 2010
Kill Switch
Toxin/antidote system
Killswitch coupled to sporulation in Bacillus subtilis
Another way of killing the cells once the biofilm has formed would be to alter an already existing in B. subtillis Within Bacillus subtilis a system exists whereby the sporulation process is delayed by killing surrounding cells. Cells that are Spo0A-Off are lysed whereby nutrients are released. These nutrients can be consumed by the Spo0A-On cells thereby delaying the process of sporulation The skf and sdp operons are responsible for the death of a subpopulation of sporulating bacterial cells. The regulatory protein Spo0A up regulates the sporulating killing factor (skfA-H operon) and the sporulating delay protein (sdpABC operon). Surrounding cells that are Spo0A-Off are killed by the extracellular killing factor skf, produced by Spo0A-On cells. SkfE and SkfF together probably act as an export pump that secretes the killing factor and thereby protect the cell. Another killing factor is sdpC. The Spo0A-On cells have two mechanisms that provide resistance to sdpC. The first one is the SdpC-SdpI-SdpR signaling pathway. The second is the repression of AbrB synthesis. A schematic overview of this mechanism is given below. If any of the factors that protect the cells from their own toxins were dysfunctional, the cells would kill themselves as soon as the Spo0A system would be turned on due nutrient limitation (which will happen inevitably in a biofilm after a while).
Anti-sense RNA regulation
In Gram-positive bacteria antisense RNA-regulated stability is found.
The so-called par region encodes for two small RNAs , RNAI and RNAII as depicted in the above figure. RNAI is 210nt long and contains the fst gene which encodes the toxic compound. RNAII is only 65nt long and highly complementary to RNAI. This results in complex formation whereby RNAI is not able to translate the fst gene into the faecalis plasmid-stabilizing toxin compound. The half-life time of RNAI is only 10 minutes while the half-life time of RNAII is more than one hour. (Zielenkiewicz)(Greenfield) This difference can be used as some sort of timer on a killswitch since the toxin concentration will eventually outcompete the antitoxin concentration. Our model is based on this kill switch system.
Promoters
To ensure a bigger time delay between chaplin expression and kill switch activation, the srfA promoter could start the chaplin expression, whereas the yqxM promoter which is active later in the growth curve would start the kill switch.
The activation of the par region can be modeled with a simple activator term. The degradation of the two RNAs can be modelled with half life equations. The change in biomass can be modeled with a simple growth model combined with
the delta tox.
The model
The formation of RNAI and RNAII is described with the Hill's equation. The degradation of the RNA is taken into account by using the half-life of the different RNA's. The Hill's equation is the same for both RNA's because the assumption is made that both promoters are equal in strength.
In the following table all constants used in the model are given.
The assumption is made that the promoter is very strong, therefore the maximal expression level of the promoter is assumed to be 1 mRNA molecule per second. Normally promoter expression levels range between 10-4 and 1 mRNA molecules per second. K is the activation coefficient and defines the concentration active X needed to significantly activate expression. The concentration of transcription factor is assumed to be much higher than the activation coefficient K. A value of 500 nM is chosen for this model.
The figure shows the amount of mRNA molecules in time. Assuming that all available mRNA II binds to mRNA I, the red line in the graph shows the amount of free mRNA I molecules. Translation of these mRNA I molecules results in the formation of a toxic compound. Above a certain threshold this will result in cell death.
Matlab source files
Killswitch ([http://dl.dropbox.com/u/391356/iGEM/killswitchMatlab.zip source])
References
Gazit E and Sauer RT, The Doc Toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. Journal of Biological Chemistry, 1999, 274 (24):16813-8. PMID 10358024
Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE, The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism, Molecular Microbiology, 2000, 37(3): 652-660. PMID 10931358
Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R, Bacterial programmed cell death and multicellular behavior in bacteria, Plos genetics 2(10): e135. PMID 15036322
Zielenkiewicz U and Ceglowski P, Mechanisms of plasmid stable maintenance with special focus on plasmid addiction ystems, Acta Biochimica Polonia, 2001, 48 (4): 1003-1023. PMID 11995964
 "
"