Team:TU Delft/Project/alkane-degradation/results
From 2010.igem.org
(→Characterization of the long-chain alkane monooxygenase; LadA) |
(→Characterization of the Alcohol DeHydrogenase (ADH) system) |
||
| Line 100: | Line 100: | ||
===[https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ADH Characterization of the Alcohol DeHydrogenase (ADH) system]=== | ===[https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ADH Characterization of the Alcohol DeHydrogenase (ADH) system]=== | ||
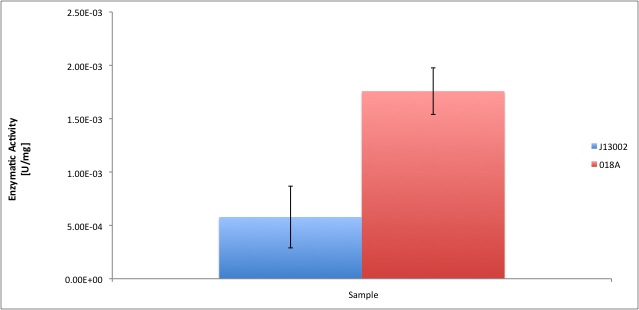
According to our results, the ''E. coli'' cell extract has a dodecanol-1 dehydrogenase activity of 9.64e-12 kat/mg (0.58 mU/mg); whereas our recombinant strain expressing the Biobrick [http://partsregistry.org/Part:BBa_K398018 BBa_K398018] has an activity of 2.93e-11 kat/mg (1.76 mU/mg), an improvement of 2-fold compared to the wild type activity; which also means 3% of the activity of the positive control ''Pseudomonas putida''. | According to our results, the ''E. coli'' cell extract has a dodecanol-1 dehydrogenase activity of 9.64e-12 kat/mg (0.58 mU/mg); whereas our recombinant strain expressing the Biobrick [http://partsregistry.org/Part:BBa_K398018 BBa_K398018] has an activity of 2.93e-11 kat/mg (1.76 mU/mg), an improvement of 2-fold compared to the wild type activity; which also means 3% of the activity of the positive control ''Pseudomonas putida''. | ||
| - | |||
| - | |||
If you are interested in knowing more about our findings, [https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ADH CLICK ON THIS LINK]. | If you are interested in knowing more about our findings, [https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ADH CLICK ON THIS LINK]. | ||
| + | |||
| + | [[Image:TUDelftADH_final.jpg|600px|thumb|center|Comparison between E. coli ADH activity and our recombinant strain. ]] | ||
===[https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ALDH Characterization of the ALdehyde DeHydrogenase system]=== | ===[https://2010.igem.org/Team:TU_Delft/Project/alkane-degradation/results/ALDH Characterization of the ALdehyde DeHydrogenase system]=== | ||
Revision as of 19:22, 27 October 2010

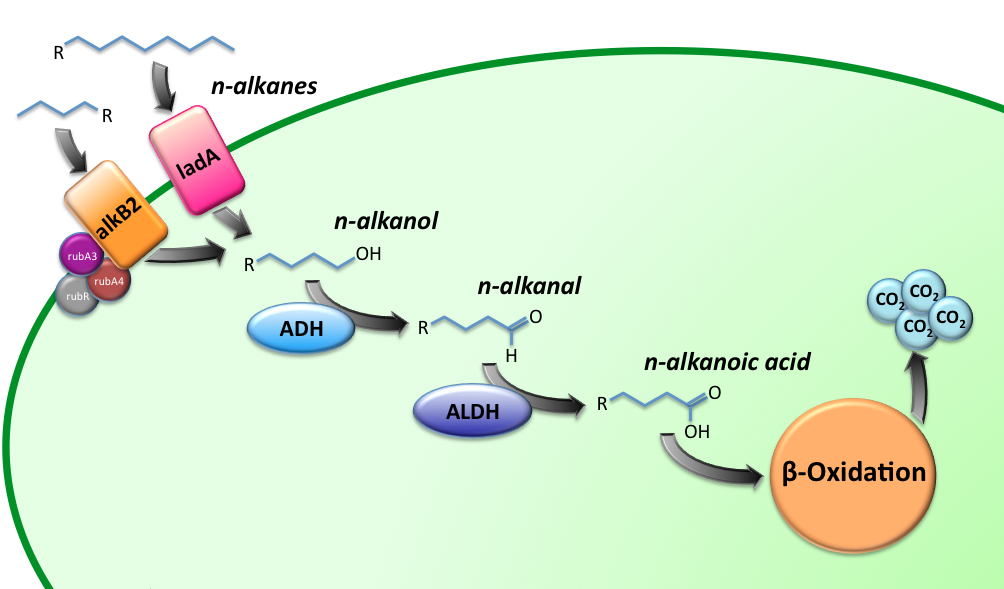
Alkane Degradation Results & Conclusions
Characterization of the alkane hydroxylase system
The following table contains the average ratio of the hexadecane/undecane surface areas. Strains numbers 1 and 2 are two positive colonies taken from the same plate. From the ratios we may conclude that the samples obtained from the E.coli strain, carrying the AH system, contain relatively less octane than the control strain. By comparing peak ratios we were able to estimate the specific enzymatic activity of the system, which was found to be 0.045 U/mg. For more information about our findings CLICK HERE.
| Strain | Ratio octane/undecane | Standard deviation [%] | Octane converted [umol] | Protein [mg] | Enzymatic activity [U/mg protein] |
| Blank | 0.832 | 6.16 | 0.00 | 0.00 | 0.00 |
| E.coli K12 negative control | 0.821 | 13.3 | 1.34 | 1.51 | 1.23E-3 |
| E.coli K12 strain #1 (AH-system) | 0.652 | 14.4 | 21.6 | 1.70 | 1.77E-2 |
| E.coli K12 strain #2 (AH-system) | 0.439 | 3.96 | 47.0 | 1.46 | 4.49E-2 |
Characterization of the long-chain alkane monooxygenase; LadA
The analysis of the GC graphs allowed us to estimate the enzymatic activity. The following table displays the average ratios found and the enzyme activity estimates. Strains numbers 1 and 2 are two positive colonies taken from the same plate.
We observed a significant increase in enzyme activity in the strains carrying the ladA protein generator compared to the negative control strain. The highest enzyme activity value was found to be 3.33E-03 U/mg protein. In order to know more about the characterization of this system, CLICK ON THIS LINK
| <b>Strain | Average ratio hexadecane/undecane | Standard deviation [%] | Hexadecane converted [umol] | Protein [mg] | Enzymatic activity [U/mg protein] |
| Blank | 0.900 | 3.19 | 0.00 | 0.00 | 0.00 |
| E.coli K12 negative control ([http://partsregistry.org/Part:BBa_J13002 J13002]) | 0.807 | 3.48 | 1.76 | 4.46 | 5.49E-04 |
| E.coli K12 strain #1 ([http://partsregistry.org/Part:BBa_K398017 K398017]) | 0.600 | 15.4 | 5.68 | 4.58 | 1.72E-03 |
| E.coli K12 strain #2 ([http://partsregistry.org/Part:BBa_K398017 K398017]) | 0.532 | 4.25 | 6.95 | 3.18 | 3.03E-03 |
| E.coli TOP10 strain ([http://partsregistry.org/Part:BBa_K398027 K398027]) | 0.492 | 2.98 | 7.71 | 3.22 | 3.33E-03 |
Characterization of the Alcohol DeHydrogenase (ADH) system
According to our results, the E. coli cell extract has a dodecanol-1 dehydrogenase activity of 9.64e-12 kat/mg (0.58 mU/mg); whereas our recombinant strain expressing the Biobrick [http://partsregistry.org/Part:BBa_K398018 BBa_K398018] has an activity of 2.93e-11 kat/mg (1.76 mU/mg), an improvement of 2-fold compared to the wild type activity; which also means 3% of the activity of the positive control Pseudomonas putida.
If you are interested in knowing more about our findings, CLICK ON THIS LINK.
Characterization of the ALdehyde DeHydrogenase system
USEFUL LITERATURE AND REFERENCES
- Kato T. et al. "Gene cloning and characterization of an aldehyde dehydrogenase from long-chain alkane-degrading Geobacillus thermoleovorans B23" Extremophiles (2010) 14:33-39.
- http://mbel.kaist.ac.kr/lab/research/protein_en1.html
- Hoffmann F. and Rinas U. "Stress Induced by Recombinant Protein Production in Escherichia coli" Advances in Biochemical Engineering/Biotechnology, 2004, Vol. 89/2004, pp. 73-92.

 "
"