Team:Stockholm/19 October 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Gel verification) |
m |
||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
| + | ==Nina== | ||
| + | |||
| + | ===Agarose gel on Colony PCR=== | ||
| + | |||
| + | I ran agarose gels on all the colony PCR screens from yesterday to check if I had inserts. | ||
| + | |||
| + | [[Image:Aq1.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq2.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq3.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq4.jpg|200px]] | ||
| + | |||
| + | Unfortunately non of these gels show that there has been an proper insert of Fusion and Protein A all with the three CPPs in the N-terminal in the pEX vector. | ||
| + | |||
| + | I made a bad decision to conduct the ligation of these parts with the pEX vector and transform the ligations directly into overexpression cells such as BL21. I should have transformed the ligations into cloning cells such as Top 10 and from there as usual perform a mini prep and transform the vectors with insert into BL21 cells. However I hoped some of the ligations that would have ended up as vectors with insert would succesfully get transformed into BL21, but it seems from the gels that this has not happened. We are short of time in the competition and this was a test to see if I could skip the two days I had to spend with transformation into Top 10, but it unfortunatelly did not work out this time. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
==Andreas== | ==Andreas== | ||
| Line 30: | Line 56: | ||
Incorrect size. Send to the Uppsala team for analysis. | Incorrect size. Send to the Uppsala team for analysis. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[Image: | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === SOD activity === | ||
| + | -continuing | ||
| + | |||
| + | |||
| + | |||
| + | *Sonicate 3x30s at ~12amp | ||
| + | |||
| + | *Make solutions | ||
| + | **WST working solution 20ml | ||
| + | **Enzyme working solution 2.5ml | ||
| + | |||
| + | |||
| + | |||
| + | {| | ||
| + | ! mix | ||
| + | | samples | ||
| + | | blank 1 | ||
| + | | blank 2 | ||
| + | | blank 3 | ||
| + | |- | ||
| + | | sample solution | ||
| + | | 20 | ||
| + | | | ||
| + | | 20 | ||
| + | | | ||
| + | |- | ||
| + | | ddH<sub>2</sub>O | ||
| + | | | ||
| + | | 20 | ||
| + | | | ||
| + | | 20 | ||
| + | |- | ||
| + | | WST solution | ||
| + | | 200 | ||
| + | | 200 | ||
| + | | 200 | ||
| + | | 200 | ||
| + | |- | ||
| + | | Enzyme solution | ||
| + | | 20 | ||
| + | | 20 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | dilution buffer | ||
| + | | | ||
| + | | | ||
| + | | 20 | ||
| + | | 20 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 240µl | ||
| + | | 240µl | ||
| + | | 240µl | ||
| + | | 240µl | ||
| + | |} | ||
| + | |||
| + | *Incubate in 37°C for 20 min | ||
| + | |||
| + | *Measure A<sub>440</sub> with nanodrop | ||
| + | |||
| + | |||
| + | *SOD activity (inhibition %) = ((A<sub>blank1</sub> - A<sub>blank3</sub>) - (A<sub>sample</sub> - A<sub>blank2</sub>))/(A<sub>blank1</sub> - A<sub>blank3</sub>) x 100 | ||
| + | |||
| + | [[Image:SOD_activity1.jpg| 900px]] | ||
| + | |||
| + | |||
| + | ==== cultures ==== | ||
| + | Top10 | ||
| + | *pEX.SOD.his.RBS.yCCS 1 | ||
| + | *pEX.SOD.his.RBS.yCCS 1 | ||
| + | BL21 | ||
| + | *pEX.SOD.RBS.yCCS | ||
| + | *pEX.his.SOD.RBS.yCCS | ||
| + | |||
| + | |||
| + | === ON cultures === | ||
| + | {| | ||
| + | | pSB1C3.nLMWP | ||
| + | | ) | ||
| + | |- | ||
| + | | pSB1C3.nTAT | ||
| + | | \ plasmid prep. | ||
| + | |- | ||
| + | | pSB1C3.nTra10 | ||
| + | | / | ||
| + | |- | ||
| + | | pEX.SOD.his.RBS.yCCS | ||
| + | | ) | ||
| + | |- | ||
| + | | pEX.SOD | ||
| + | | \ SOD activity test | ||
| + | |- | ||
| + | | pEX.yCCS | ||
| + | | / | ||
| + | |- | ||
| + | | pEX.bFGF | ||
| + | | ) | ||
| + | |- | ||
| + | | pEX.IgGp | ||
| + | | / | ||
| + | |- | ||
| + | | pEX.Prot.A | ||
| + | | > glycerol stock | ||
| + | |- | ||
| + | | pEX.SOD | ||
| + | | \ | ||
| + | |- | ||
| + | | pEX.yCCS | ||
| + | | ) | ||
| + | |} | ||
| + | |||
| + | =Johan= | ||
| + | |||
| + | ==Miniprep== | ||
| + | 3 ml of sampled "5-6", "5-8", "AS10" and "EA3". Verification of the last constructs, his-bFGF, bFGF-his and tat-bFGF-his. | ||
| + | |||
| + | ==Cut miniprep== | ||
| + | |||
| + | 5 µl DNA | ||
| + | |||
| + | (1 µl BamHI enzyme) | ||
| + | |||
| + | 2 µl 10x buffer | ||
| + | |||
| + | 12 µl H2O | ||
| + | |||
| + | ==PCR screen== | ||
| + | Of the other constructs in BL21 cells, tra10-bFGF-his, lmwp-bfgf-his, his-bFGF-tra10, his-bFGF-tat, his-bFGF-lmwp. 2 colonies for each plate. | ||
| + | |||
| + | 0,5 µl Taq pol | ||
| + | |||
| + | 0,5 µl dNTP | ||
| + | |||
| + | 5 µl 5x buffer | ||
| + | |||
| + | 1 µl pex for primer | ||
| + | |||
| + | 1 µl pex rev primer | ||
| + | |||
| + | 17 µl H2O | ||
| + | |||
| + | A mastermix 10x was made | ||
| + | |||
| + | ==Gel of minipreps and PCR screen== | ||
| + | |||
| + | Top lanes: Cut & uncut miniprep | ||
| + | |||
| + | Bottom lanes: PCR screens | ||
| + | |||
| + | [[Image:SU 19oktgel.png]] | ||
| + | |||
| + | Results: | ||
| + | |||
| + | Top lanes, I realized I put BamHI even in the samples that was supposed to be uncut! | ||
| + | |||
| + | Bottom lanes: Correct size in the first lane but otherwise empty, why? | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 19:03, 27 October 2010
Contents |
Nina
Agarose gel on Colony PCR
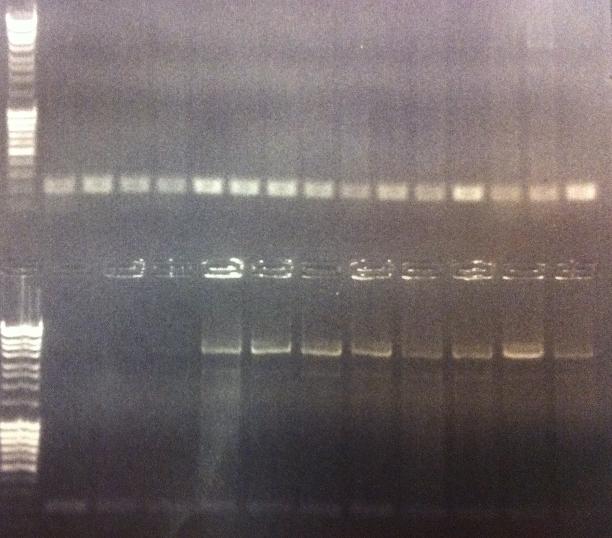
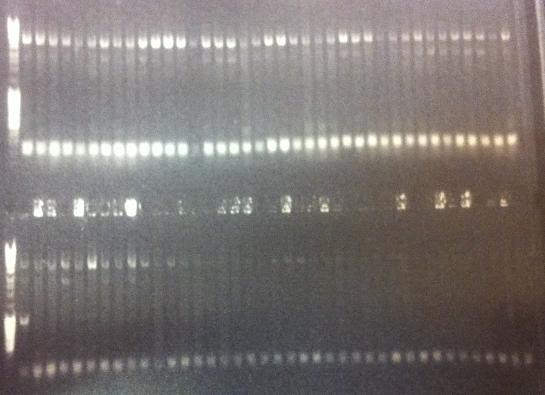
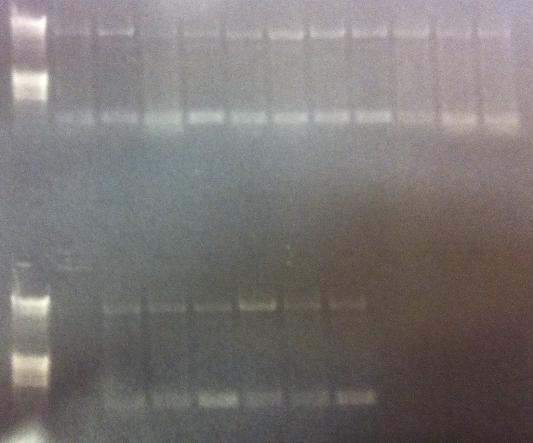
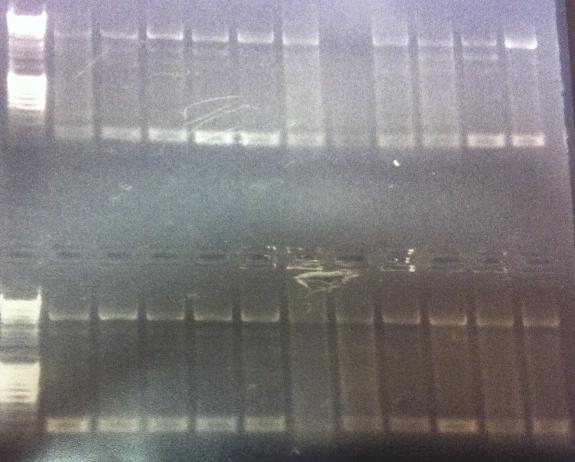
I ran agarose gels on all the colony PCR screens from yesterday to check if I had inserts.
Unfortunately non of these gels show that there has been an proper insert of Fusion and Protein A all with the three CPPs in the N-terminal in the pEX vector.
I made a bad decision to conduct the ligation of these parts with the pEX vector and transform the ligations directly into overexpression cells such as BL21. I should have transformed the ligations into cloning cells such as Top 10 and from there as usual perform a mini prep and transform the vectors with insert into BL21 cells. However I hoped some of the ligations that would have ended up as vectors with insert would succesfully get transformed into BL21, but it seems from the gels that this has not happened. We are short of time in the competition and this was a test to see if I could skip the two days I had to spend with transformation into Top 10, but it unfortunatelly did not work out this time.
Andreas
Colony PCRs
- pEX.ProtA⋅His: PA 1 & 2
- pEX.IgGp: Ig 1 & 2
Standard colony PCR settings
- Elongation time: 1:20
Gel verification
Expected bands
- PA: 417 bp
- Ig: 1149 bp
Results
All four constructs confirmed.
PCR for Uppsala team
Gel verification
Expected bands
- All constructs: ≈ 6570 bp
Results
Incorrect size. Send to the Uppsala team for analysis.
Mimmi
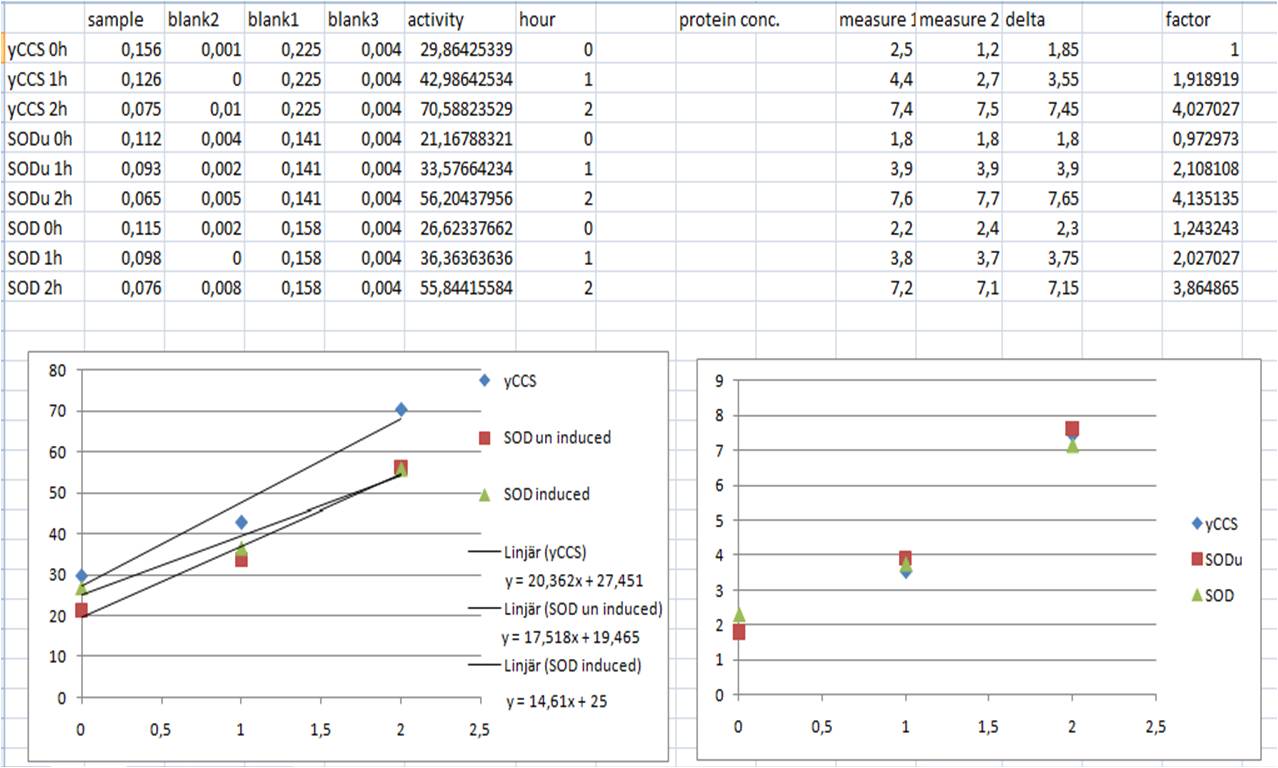
SOD activity
-continuing
- Sonicate 3x30s at ~12amp
- Make solutions
- WST working solution 20ml
- Enzyme working solution 2.5ml
| mix | samples | blank 1 | blank 2 | blank 3 |
|---|---|---|---|---|
| sample solution | 20 | 20 | ||
| ddH2O | 20 | 20 | ||
| WST solution | 200 | 200 | 200 | 200 |
| Enzyme solution | 20 | 20 | ||
| dilution buffer | 20 | 20 | ||
| tot | 240µl | 240µl | 240µl | 240µl |
- Incubate in 37°C for 20 min
- Measure A440 with nanodrop
- SOD activity (inhibition %) = ((Ablank1 - Ablank3) - (Asample - Ablank2))/(Ablank1 - Ablank3) x 100
cultures
Top10
- pEX.SOD.his.RBS.yCCS 1
- pEX.SOD.his.RBS.yCCS 1
BL21
- pEX.SOD.RBS.yCCS
- pEX.his.SOD.RBS.yCCS
ON cultures
| pSB1C3.nLMWP | ) |
| pSB1C3.nTAT | \ plasmid prep. |
| pSB1C3.nTra10 | / |
| pEX.SOD.his.RBS.yCCS | ) |
| pEX.SOD | \ SOD activity test |
| pEX.yCCS | / |
| pEX.bFGF | ) |
| pEX.IgGp | / |
| pEX.Prot.A | > glycerol stock |
| pEX.SOD | \ |
| pEX.yCCS | ) |
Johan
Miniprep
3 ml of sampled "5-6", "5-8", "AS10" and "EA3". Verification of the last constructs, his-bFGF, bFGF-his and tat-bFGF-his.
Cut miniprep
5 µl DNA
(1 µl BamHI enzyme)
2 µl 10x buffer
12 µl H2O
PCR screen
Of the other constructs in BL21 cells, tra10-bFGF-his, lmwp-bfgf-his, his-bFGF-tra10, his-bFGF-tat, his-bFGF-lmwp. 2 colonies for each plate.
0,5 µl Taq pol
0,5 µl dNTP
5 µl 5x buffer
1 µl pex for primer
1 µl pex rev primer
17 µl H2O
A mastermix 10x was made
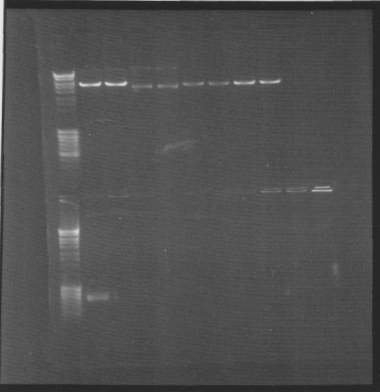
Gel of minipreps and PCR screen
Top lanes: Cut & uncut miniprep
Bottom lanes: PCR screens
Results:
Top lanes, I realized I put BamHI even in the samples that was supposed to be uncut!
Bottom lanes: Correct size in the first lane but otherwise empty, why?
 |

|
 |

|
 |

|

|

|
 "
"