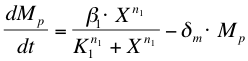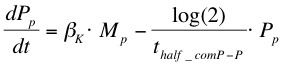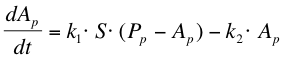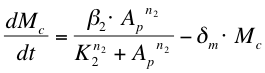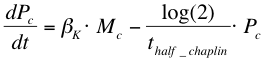Team:Groningen/Expression model
From 2010.igem.org
(→Expression) |
(→Expression) |
||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
== Expression == | == Expression == | ||
<html> | <html> | ||
| - | <img src="https://static.igem.org/mediawiki/2010/ | + | <img src="https://static.igem.org/mediawiki/2010/8/84/Groningen-pathway.png" style="float: right; width: 360px" alt="Schematic Overview" title=""> |
</html> | </html> | ||
| - | [[Image: | + | |
| + | === Introduction === | ||
| + | To form a biofilm with hydrophobic surface properties, expression of the hydrophobic proteins(in our case chaplins) should coincide with the late stages of biofilm formation. There are several reasons for this: mainly because not all bacteria participate in biofilm formation, there is significant cell differentiation prior to the formation of a biofilm. Also the expression of chaplins should not interfere with the growth speed and biofilm formation. The latter could be problematic due to the hydrophobic properties of the chaplins which might interfere with the formation of the extracellular matrix(ECM). We found that the ComXPA quorum sensing system and related gene expressions might suitable for this purpose. | ||
| + | |||
| + | |||
| + | ===ComXPA quorum sensing system=== | ||
| + | Quorum sensing in bacteria means that bacteria ‘talk’ to each other via signaling molecules. These molecules accumulate extracellularly as cultures grow to high cell densities. The response to these signaling molecules can result in dramatic changes in gene expression. In Bacillus subtilis a quorum sensing pathway named ComX-ComP-ComA is present which regulates the development of genetic competence. The ComX pheromone is a 10-amino-acid peptide which is secreted. | ||
| + | |||
| + | When ComX accumulates extracellularly, it stimulates the activity of a membrane bound receptor histidine kinase called ComP. ComP then autophosphorylates and activates the response regulator ComA by donating a phosphate molecule. The activated ComA functions as a transcriptional activator and activates the expression of 20 genes directly and the expression of another 150 genes is affected indirectly. The affected genes seem to have three different functions: the coordination of physiological changes involved in developmental pathways, the production of extracellular products and the enhancement of survival, growth and colonization. ComA directly activates the srfA operon which encodes enzymes needed for the production of the lipopeptide surfactin. The binding site for the activated ComA is known, the consensus sequence is a 12 bp palingdrome with a 4-5 bp spacer (TTGCGGNNNNCCGCAA). A DNA microarray study determined which genes were mostly activated by the active ComA (Comella et. al., 2005) . For our project we chose to link our chaplins to the promoter of the srfA gene which was most activated, which was srfAA. | ||
| + | |||
| + | |||
| + | === The model === | ||
| + | Our model describes the activation of ComP by ComX to the eventual formation of our chaplins. | ||
| + | |||
| + | The activation of ComP-P expression by ComX (''X'') is modelled with a Hill’s equation. The degradation of the formed mRNA is taken into account. The equation describes the formation of ComP-P mRNA (''Mp'') in time. | ||
| + | |||
| + | [[Image:RUG Eq_ComP-P.png]] | ||
| + | |||
| + | After the formation of mRNA, the protein ComP-P (''Pp'') will be formed which is modelled with the following equation: | ||
| + | |||
| + | [[Image:RUG Eq_P_ComP-P.png]] | ||
| + | |||
| + | The next reaction within the metabolism is the phosphorylation of ComA by ComP-P. The formation of ComA-P (''Ap'') is described by the following equation: | ||
| + | |||
| + | [[Image:RUG Eq_Phospho.png]] | ||
| + | |||
| + | After phosphorylation, ComA-P is able to activate the expression of the gene that is behind the srfA promoter. This will be one of the chaplins in our case or GFP to test the system. Again, the Hill’s equation is used to model this process combined with the degradation of mRNA and the formation of the protein. The first equation shows the formation of mRNA of chaplins (''Mc''), the second equation describes the formation of the chaplin (''Pc'') | ||
| + | |||
| + | [[Image:RUG Eq_ComAP.png]] | ||
| + | |||
| + | [[Image:RUG Eq_P_ComAP.png]] | ||
| + | |||
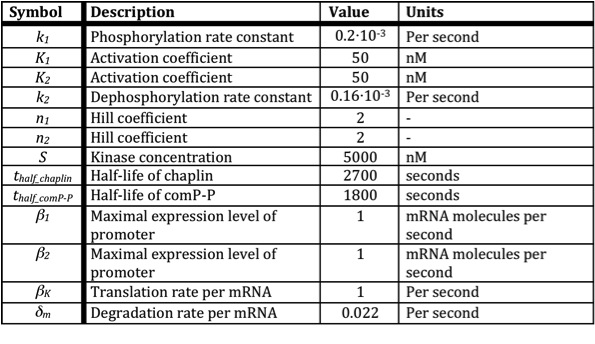
| + | Below a table is shown with all the values of the used constants. | ||
| + | |||
| + | [[Image:RUG Table_Expression.jpg]] | ||
| + | |||
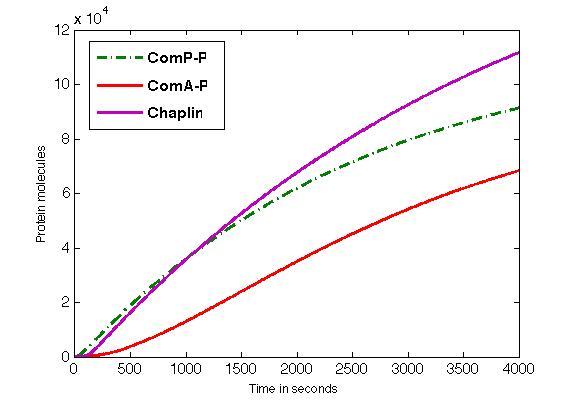
| + | The figure shows the formation of ComP-P, ComA-P and chaplins in time. | ||
| + | |||
| + | [[Image:RUG Fig_chaplin.jpg]] | ||
| + | |||
| + | |||
| + | === Matlab source files === | ||
| + | * Expression ([http://dl.dropbox.com/u/391356/iGEM/expressionMatlab.zip source]) | ||
| + | |||
| + | |||
| + | === References === | ||
| + | <small>Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP, Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay, Proceedings of the National Academy of Sciences, 2009, 106 (16): 6459-64. PMID 19380751 (activationcoefficient K) | ||
| + | |||
| + | Mitrophanov AY, Groisman EA, Positive feedback in cellular control systems, Bioessays, 2008, (6): 542-55. PMID 18478531 (Hills factor) | ||
| + | |||
| + | Comella N, Grossman AD., Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis, Molecular Microbiology (2005) 57 (4), 1159–1174. PMID 16091051 | ||
| + | |||
| + | Leisner M, Kuhr JT, Rädler JO, Frey E, Maier B, Kinetics of genetic switching into the state of bacterial competence, Biophysical Journal, 2009, 96(3): 1178-88. PMID 19186153 | ||
| + | |||
| + | Leisner M, Stingl K, Frey E, Maier B, Stochastic switching to competence, Current Opinion in Microbiology, 2008, 11(6):553-9. PMID 18955155 | ||
| + | |||
| + | Leisner M, Stingl K, Rädler JO, Maier B, Basal expression rate of comK sets a 'switching-window' into the K-state of Bacillus subtilis, Molecular Microbiology, 2007, 63 (6): 1806-16. PMID 17367397 | ||
| + | |||
| + | Najafi SM, Harris DA, Yudkin MD, Properties of the phosphorylation reaction catalyzed by SpoIIAB that help to regulate sporulation of Bacillus subtilis, Journal of Bacteriology, 1997, 179 (17): 5628-5631. PMID 9287028 (phosphorylation rates and kinase concentration) | ||
| + | |||
| + | Roggiani M, Dubnau D., ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA, Journal of Bacteriology 1993 175(10):3182-7. PMID 8387999 | ||
| + | |||
| + | Shah IM, Dworkin J., Microbial interactions: bacteria talk to (some of) their neighbors, Current Biology, 2009 19(16): 689-91. PMID 19706277 | ||
| + | |||
| + | Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB, An excitable gene regulatory circuit induces transient cellular differentiation, Nature, 2006, 440 (7083): 545-50. PMID 16554821 | ||
| + | </small> | ||
Latest revision as of 18:43, 27 October 2010
Expression

Introduction
To form a biofilm with hydrophobic surface properties, expression of the hydrophobic proteins(in our case chaplins) should coincide with the late stages of biofilm formation. There are several reasons for this: mainly because not all bacteria participate in biofilm formation, there is significant cell differentiation prior to the formation of a biofilm. Also the expression of chaplins should not interfere with the growth speed and biofilm formation. The latter could be problematic due to the hydrophobic properties of the chaplins which might interfere with the formation of the extracellular matrix(ECM). We found that the ComXPA quorum sensing system and related gene expressions might suitable for this purpose.
ComXPA quorum sensing system
Quorum sensing in bacteria means that bacteria ‘talk’ to each other via signaling molecules. These molecules accumulate extracellularly as cultures grow to high cell densities. The response to these signaling molecules can result in dramatic changes in gene expression. In Bacillus subtilis a quorum sensing pathway named ComX-ComP-ComA is present which regulates the development of genetic competence. The ComX pheromone is a 10-amino-acid peptide which is secreted.
When ComX accumulates extracellularly, it stimulates the activity of a membrane bound receptor histidine kinase called ComP. ComP then autophosphorylates and activates the response regulator ComA by donating a phosphate molecule. The activated ComA functions as a transcriptional activator and activates the expression of 20 genes directly and the expression of another 150 genes is affected indirectly. The affected genes seem to have three different functions: the coordination of physiological changes involved in developmental pathways, the production of extracellular products and the enhancement of survival, growth and colonization. ComA directly activates the srfA operon which encodes enzymes needed for the production of the lipopeptide surfactin. The binding site for the activated ComA is known, the consensus sequence is a 12 bp palingdrome with a 4-5 bp spacer (TTGCGGNNNNCCGCAA). A DNA microarray study determined which genes were mostly activated by the active ComA (Comella et. al., 2005) . For our project we chose to link our chaplins to the promoter of the srfA gene which was most activated, which was srfAA.
The model
Our model describes the activation of ComP by ComX to the eventual formation of our chaplins.
The activation of ComP-P expression by ComX (X) is modelled with a Hill’s equation. The degradation of the formed mRNA is taken into account. The equation describes the formation of ComP-P mRNA (Mp) in time.
After the formation of mRNA, the protein ComP-P (Pp) will be formed which is modelled with the following equation:
The next reaction within the metabolism is the phosphorylation of ComA by ComP-P. The formation of ComA-P (Ap) is described by the following equation:
After phosphorylation, ComA-P is able to activate the expression of the gene that is behind the srfA promoter. This will be one of the chaplins in our case or GFP to test the system. Again, the Hill’s equation is used to model this process combined with the degradation of mRNA and the formation of the protein. The first equation shows the formation of mRNA of chaplins (Mc), the second equation describes the formation of the chaplin (Pc)
Below a table is shown with all the values of the used constants.
The figure shows the formation of ComP-P, ComA-P and chaplins in time.
Matlab source files
- Expression ([http://dl.dropbox.com/u/391356/iGEM/expressionMatlab.zip source])
References
Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP, Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay, Proceedings of the National Academy of Sciences, 2009, 106 (16): 6459-64. PMID 19380751 (activationcoefficient K)
Mitrophanov AY, Groisman EA, Positive feedback in cellular control systems, Bioessays, 2008, (6): 542-55. PMID 18478531 (Hills factor)
Comella N, Grossman AD., Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis, Molecular Microbiology (2005) 57 (4), 1159–1174. PMID 16091051
Leisner M, Kuhr JT, Rädler JO, Frey E, Maier B, Kinetics of genetic switching into the state of bacterial competence, Biophysical Journal, 2009, 96(3): 1178-88. PMID 19186153
Leisner M, Stingl K, Frey E, Maier B, Stochastic switching to competence, Current Opinion in Microbiology, 2008, 11(6):553-9. PMID 18955155
Leisner M, Stingl K, Rädler JO, Maier B, Basal expression rate of comK sets a 'switching-window' into the K-state of Bacillus subtilis, Molecular Microbiology, 2007, 63 (6): 1806-16. PMID 17367397
Najafi SM, Harris DA, Yudkin MD, Properties of the phosphorylation reaction catalyzed by SpoIIAB that help to regulate sporulation of Bacillus subtilis, Journal of Bacteriology, 1997, 179 (17): 5628-5631. PMID 9287028 (phosphorylation rates and kinase concentration)
Roggiani M, Dubnau D., ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA, Journal of Bacteriology 1993 175(10):3182-7. PMID 8387999
Shah IM, Dworkin J., Microbial interactions: bacteria talk to (some of) their neighbors, Current Biology, 2009 19(16): 689-91. PMID 19706277
Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB, An excitable gene regulatory circuit induces transient cellular differentiation, Nature, 2006, 440 (7083): 545-50. PMID 16554821
 "
"