HKU-Hong Kong/12 July 2010
From 2010.igem.org
| Line 1: | Line 1: | ||
| + | {{:Team:HKU-Hong_Kong/header}} | ||
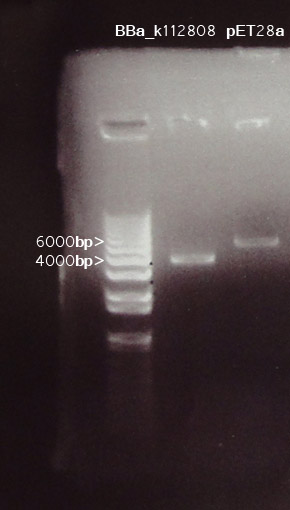
[[Image:12jul.jpg|300px|frame|double enzyme digestion of BBa_k112808 & pET28a by NotI &XbaI]] | [[Image:12jul.jpg|300px|frame|double enzyme digestion of BBa_k112808 & pET28a by NotI &XbaI]] | ||
<h1>Our official start:12 July 2010</h1> | <h1>Our official start:12 July 2010</h1> | ||
| Line 27: | Line 28: | ||
[[Team:HKU-Hong_Kong|Home]] | [[Team:HKU-Hong_Kong|Home]] | ||
| + | {{:Team:HKU-Hong_Kong/footer}} | ||
Revision as of 17:59, 27 October 2010

Contents |
Our official start:12 July 2010
After 2 weeks of training on basic techniques on biological lab work, we started our project by testing the functionality of the suicide gene.
Background
The biopart BBa_k112808 shipped is in the plasmid backbone pSB1A2. We tried to separate the biopart from the plasmid by enzyme digestion. After looking into the sequences, double enzyme digestion with NotI and XbaI was carried out.
Aim
To obtain biopart BBa_k112808 for further modifications and combination with other bioparts in different plasmid.
Results
A gel photo was taken for detection of cut DNA fragments.
BBa_k112808
Two bands are expected for the enzyme digestion on BBa_k112808. Yet, a band with ~4000bp was observed. This size matches with the SUM of the two fragments expected.
BBa_k112808:~1.8k bp
pSB1A2: ~1.9k bp
Some of the restriction sites might be absent for such a result to be obtained.
pET28a
The size of pET28a is 5369 bp, with reference to its genome map, the two fragemnts were expected to be ~5000bp and ~200bp. As a 1% agarose gel was used, one band at ~5000bp was expected. Consistence was observed in the digestion.
 "
"