Team:Freiburg Bioware/Project/Results/ITRDiary
From 2010.igem.org
| Line 1: | Line 1: | ||
| + | {{:Team:Freiburg_Bioware/Head}}{{:Team:Freiburg_Bioware/jquery}}{{:Team:Freiburg_Bioware/menu_home}} | ||
| + | |||
<p><span style="font-family:Segoe Script; font-size:25px; font-weight:bold;">Dear Diary,</span><br> | <p><span style="font-family:Segoe Script; font-size:25px; font-weight:bold;">Dear Diary,</span><br> | ||
<br/> | <br/> | ||
| Line 193: | Line 195: | ||
*Srivastava, a, Lusby, E.W. & Berns, K I, 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. Journal of virology, 45(2), pp.555-64. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=256449&tool=pmcentrez&rendertype=abstract. | *Srivastava, a, Lusby, E.W. & Berns, K I, 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. Journal of virology, 45(2), pp.555-64. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=256449&tool=pmcentrez&rendertype=abstract. | ||
<br/> | <br/> | ||
| + | {{:Team:Freiburg_Bioware/Footer}} | ||
Latest revision as of 17:48, 27 October 2010
Dear Diary,
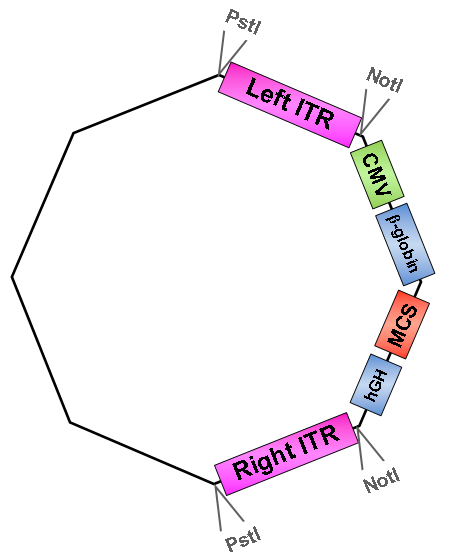
once upon a time there were two inverted terminal repeats (ITRs) – the left and the right ITR. They flanked a CMV promoter, followed by a β-globin intron, a multiple cloning site and a hGH polyA site in order to be packaged into Adeno-associated virus serotype 2 particles.
One day some biology students, participating at the iGEM competition, wanted to produce BioBricks of these two DNA sequences.
26.05.2010
For this purpose they designed and ordered primers in order to delete the PstI restriction sites which flank the ITRs via site-directed mutagenesis.
02.06.2010
Primers for removal of NotI restriction sites, which also flank the ITRs, were designed and ordered.
10.06.2010
The students conducted the first attempt of removing the PstI restriction sites: The 1st PCR for site-directed mutagenesis was performed…
… but didn’t yield any results.
18.06.2010
The team encountered a problem: The designed primers bound to both ITRs, thus amplification of just one ITR was not possible. So they hit upon the idea of digesting the plasmid with AlwNI in order to produce two different-sized fragments. Prior to PCR these two sequences could be separated by electrophoresis.
25.06.2010
Plasmid was digested with AlwNI.
28.06.2010
The 2nd site-directed mutagenesis PCR was performed…
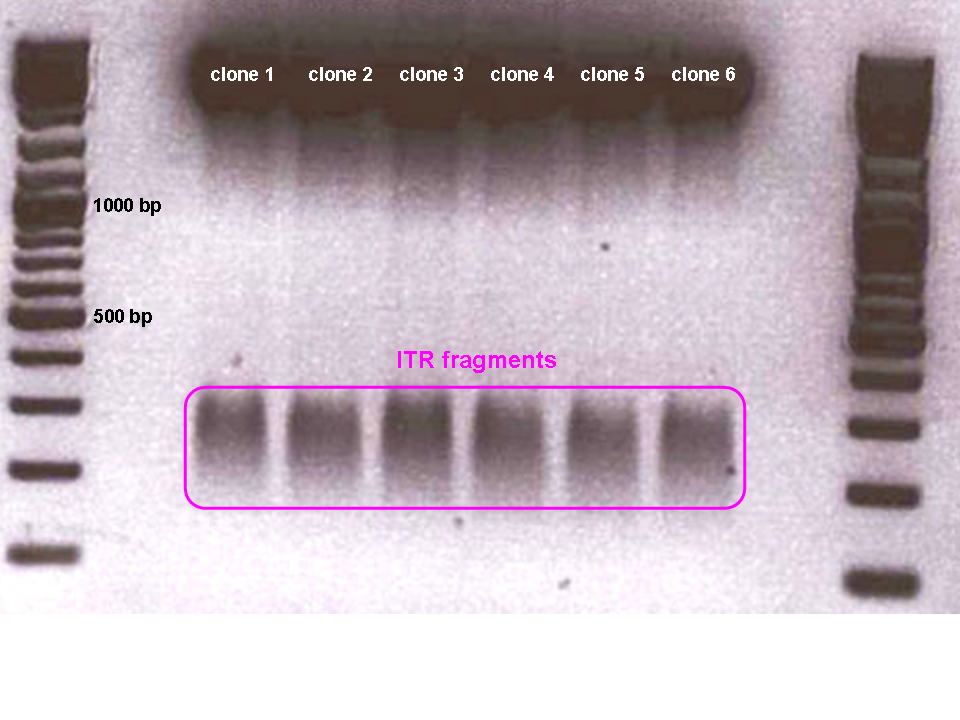
… but again didn’t work - probably due to GC-rich regions which result in the very strong secondary structure of the inverted terminal repeats (see left picture :) ).
The students didn’t burry their head in the sand: New primers with higher annealing temperatures were designed and ordered.
30.06.2010
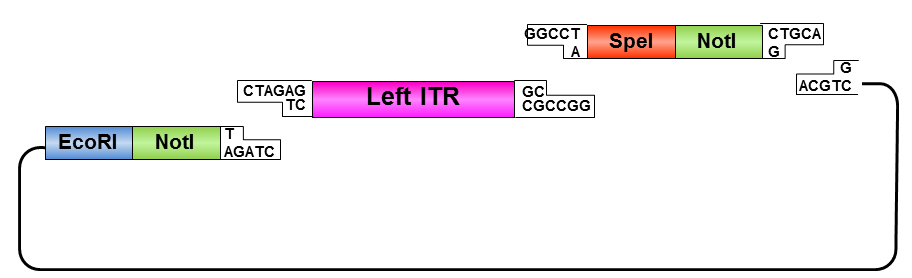
By providing RFC10 restriction sites the ITR_oligo construct could be cloned into the iGEM standard plasmid resulting in ITR BioBrick. The FreiGEMs named this alternative plan the “ITR fancy method”.
05.07.2010
The “ITR fancy method” was specified:
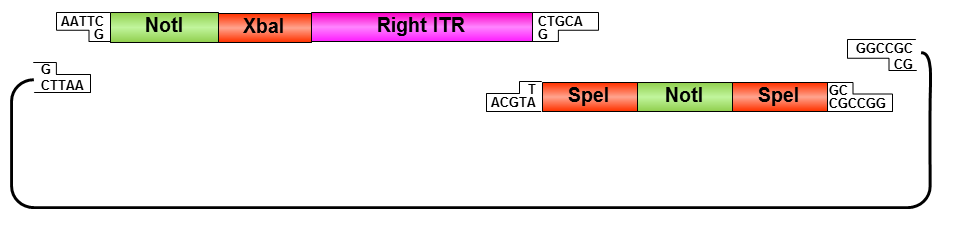
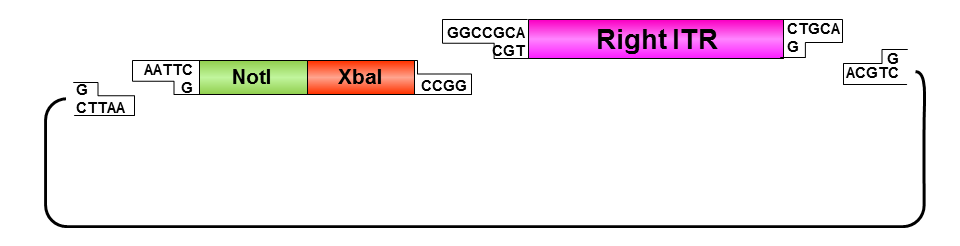
Oligos were designed which needed to be hybridized and provided necessary RFC10 prefix or suffix restriction sites. They possesed compatible overhangs to the PstI and NotI restriction sites of the ITRs but shouldn’t generate new restriction sites after ligation.
ITR fancy method:
• Digestion of pAAV_MCS with AlwNI: results in one large fragment (2982 bp) containing the right ITR and one small fragment (1674 bp) containing the left ITR.
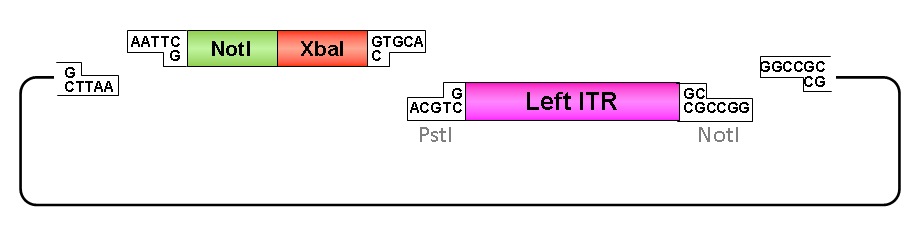
Left ITR:
• Digestion of left ITR fragment with PstI and NotI
• Digestion of pSB1C3 with EcoRI and NotI
• Ligation of left ITR with prefix and pSB1C3
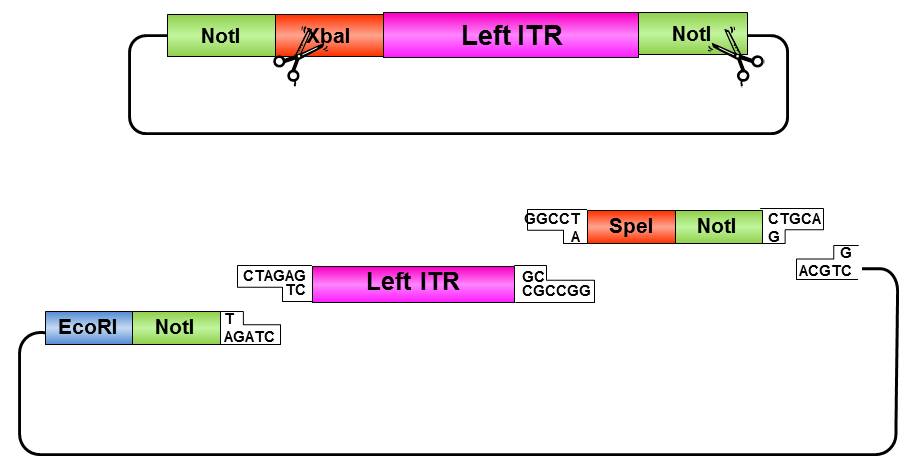
• Digestion of pSB1C3_prefix_leftITR with XbaI and NotI
• Digestion of pSB1C3 with PstI and XbaI
• Ligation of left ITR with suffix and pSB1C3
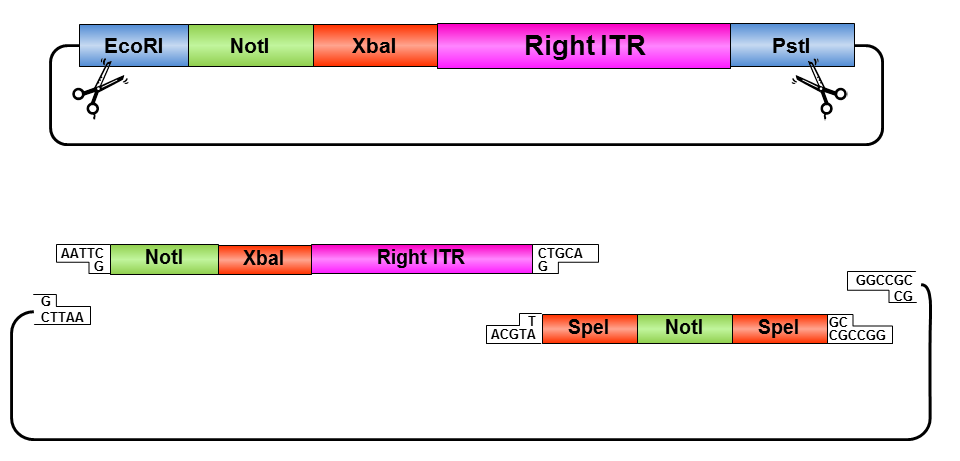
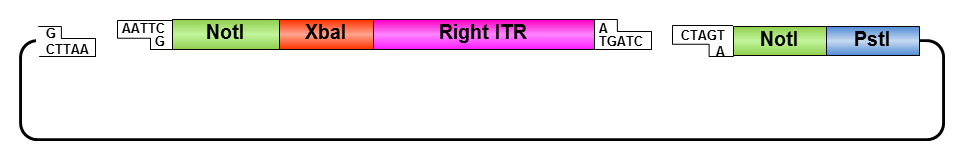
Right ITR:
• Digestion of right ITR fragment with NotI and PstI
• Digestion of pSB1C3 with XbaI and PstI
• Ligation of right ITR with prefix and pSB1C3
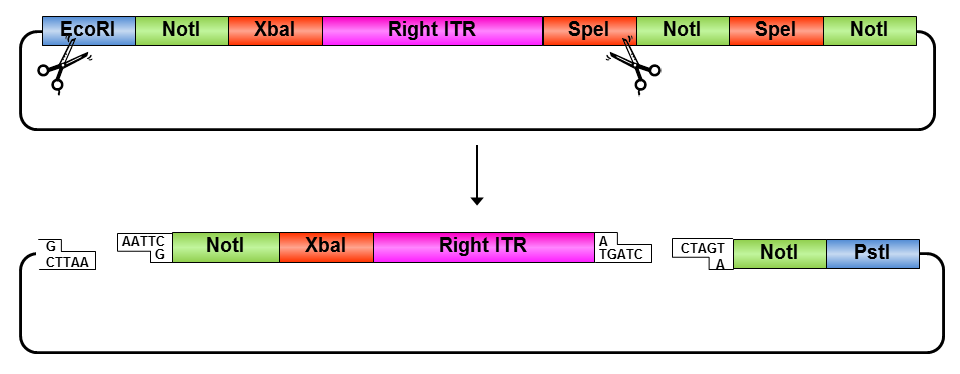
• Digestion of pSB1C3_rightITR with EcoRI and NotI
• Digestion of pSB1C3 with EcoRI and NotI
• Ligation of right ITR with special suffix and pSB1C3
• Digestion of pSB1C3_prefix_rightITR_specialsuffix with EcoRI and SpeI
• Digestion of pSB1C3 with EcoRI and SpeI
• Ligation of pSB1C3 with right ITR
On the same day the 3rd PCR attempt with new primers was conducted. Two reactions per ITR were performed – one without and one with 10% DMSO… but again no PCR product was detectable.
12.07.2010
First steps of the “ITR fancy method” were performed.
14.07.2010
The ambitious students wanted to play save and also ordered the left ITR according to the RFC10 standard at a gene synthesis company. Nevertheless they continued with PCR and cloning, because they knew by experience that DNA synthesis could possibly take long.
15.07.2010
Unfortunately it turned out that the first try of the “ITR fancy method” didn’t work. The students were not surprised: Digestion of plasmid, gel extraction, followed by second digestion and gel extraction of just 150 bp small ITR fragments leaves nearly no DNA for ligation. Nevertheless cloning was repeated.
17.07.2010
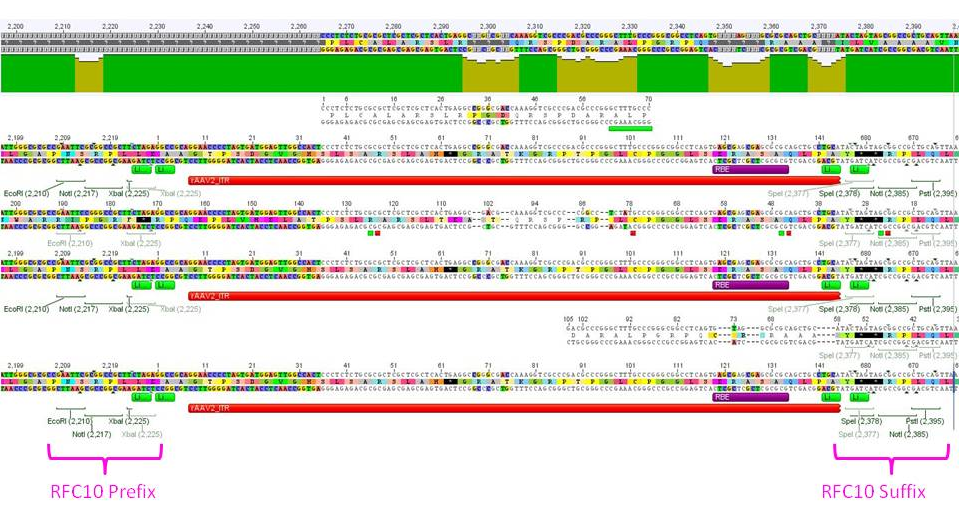
Test digestion revealed:
RFC10 prefix was successfully fused to the left ITR.
19.07.2010
Test digested of the right ITR also revealed positive results: Right ITR was provided with RFC10 prefix.
Some team impressions:
“Yess, that’s great! Finally something worked with our ITRs!” (Bea)
“The most astonishing test digestion gel of our entire working time!” (Adrian)
21.07.2010
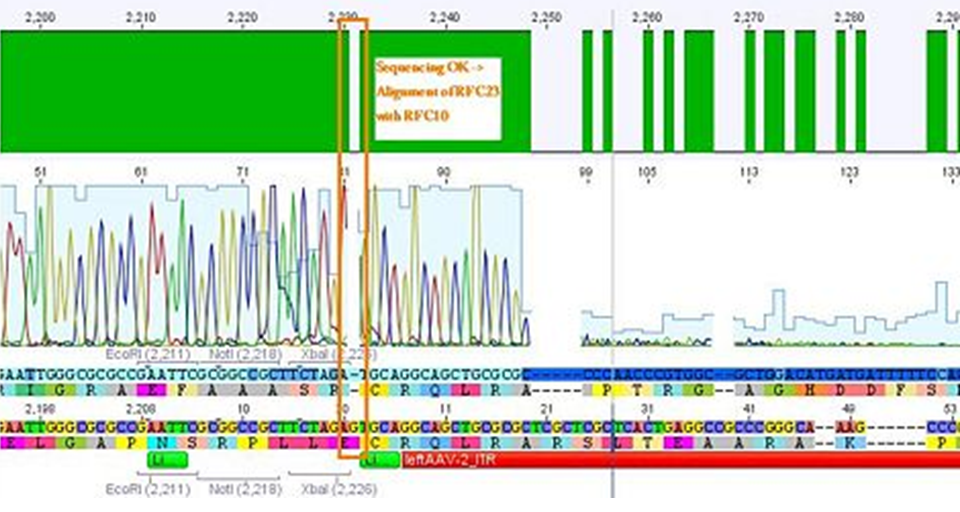
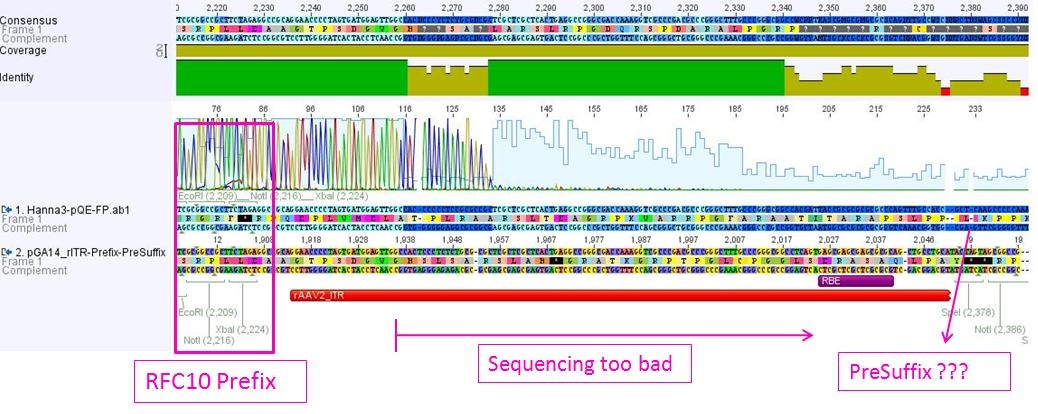
Sequencing not only revealed that the prefixes were successfully fused to the ITRs, but also that besides site-directed mutagenesis also sequencing of the ITRs following the standard procedure does not work.
The students concluded that the polymerase isn’t able to read through and that their ITRs must fulfil a great job in forming very strong secondary structures!
It turned out that a “G” was missing in the prefix of the left ITR. Therefore new oligos were designed and ordered.
26.07.2010
The left ITR fancy method was started again with the newly arrived oligos.
In addition to that cloning of the right ITR was continued: Fusion of special suffix to right ITR.
29.07.2010
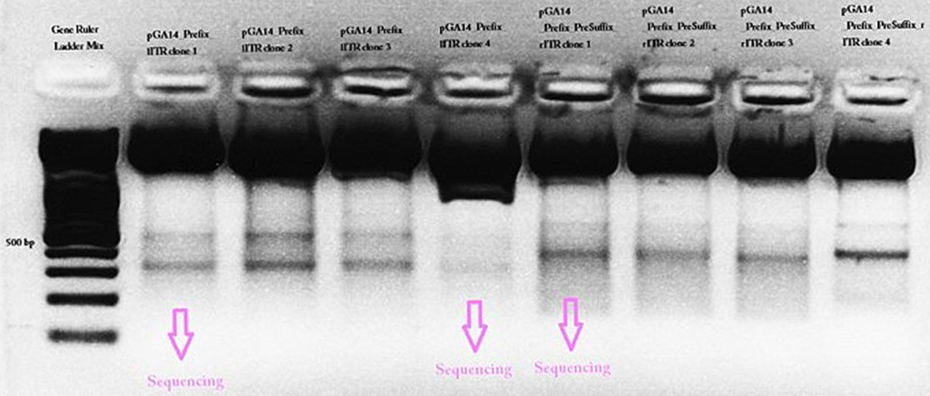
pGA14_newPrefix_leftITR and pGA14_prefix_rightITR_specialSuffix was prepared and test digested. The ITR bands didn’t exactly match to the expected fragment size. In addition a second band could be detected at about 500 bp. The students weren’t able to explain this and sent three samples for sequencing.
31.07.2010
Left ITR:
The sequencing company didn’t manage to read through the left ITR samples until now. The FreiGEMs interpreted this as good news: Problems with sequencing indicated very strong secondary structures – caused by (for example) ITRs! :)
Therefore “ITR fancy method” was continued: Suffix was fused to left ITR.
Rigth ITR:
Sequencing results confirmed test digestion: The right ITR was provided with RFC10 prefix. However due to short chromatogram range it was not possible to validate that the “special” suffix was fused to the ITR.
The students didn’t bounce back and continued with the last step of their ITR fancy method:
The right ITR needed to be subcloned into a plasmid providing the iGEM restriction sites, resulting in RFC10 suffix downstream of the ITR.
02.08.2010
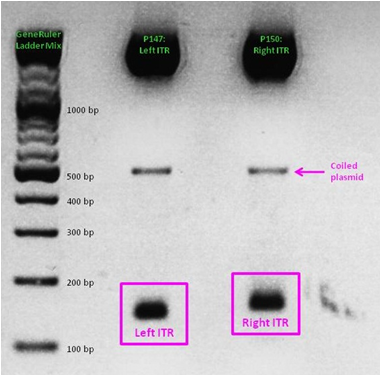
Last mini-prep and test digestion was conducted: The expected fragment sizes (156 resp. 157) could be detected in the referring range between 100 and 200 bp.
Now the students wanted to verify these results and sent the samples for sequencing – this time following special procedures.
04.08.2010
Eight sequencing files (which needed to be analyzed base by base "per hand") later, alignments showed that the ITRs are OK and were successfully converted into the RFC10 BioBrick standard.
After subcloning the ITRs into the iGEM pSB1C3 plasmid, the students were happy and proud of their success.
So they all lived happily ever after and had much fun with their new ITR BioBricks.
By the way: The students obtained satisfaction, because the ordered rebellious ITRs didn’t arrive until now – even DNA synthesis seems to be difficult….
I guess the lesson is clear: Never perform PCR with ITRs, nor try to order them from gene synthesis companies. And: Better do not try to sequence them! ;)
Good luck!
Source
- Berns, K., 1990. Parvovirus replication. Microbiology and Molecular Biology Reviews, 54(3), p.316. Available at: http://mmbr.asm.org/cgi/content/abstract/54/3/316.
- Srivastava, a, Lusby, E.W. & Berns, K I, 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. Journal of virology, 45(2), pp.555-64. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=256449&tool=pmcentrez&rendertype=abstract.
 "
"