Team:Michigan/Project
From 2010.igem.org
Chillywings (Talk | contribs) (→References) |
|||
| (3 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
The Hy-Bi project is in association with the Chemical Engineering department’s joint project to produce hydrocarbons from algae biomass, dubbed "Hy-Bi"[1,2]. This consists of hydrothermal processing of the algae followed by catalytic upgrading of the “bio crude” oil. Before the hydrothermal processing can take place the algae needs to be concentrated from 1 to 10 g/L to over 250 g/L. Traditional methods that are used like centrifugation, filtration and chemical flocculation are costly. | The Hy-Bi project is in association with the Chemical Engineering department’s joint project to produce hydrocarbons from algae biomass, dubbed "Hy-Bi"[1,2]. This consists of hydrothermal processing of the algae followed by catalytic upgrading of the “bio crude” oil. Before the hydrothermal processing can take place the algae needs to be concentrated from 1 to 10 g/L to over 250 g/L. Traditional methods that are used like centrifugation, filtration and chemical flocculation are costly. | ||
| - | For this project, we examined methods of creating a bioflocculant [ | + | For this project, we examined methods of creating a bioflocculant [3]. We will be over-expressing type I pili and their associated adhesion protein, making a hyper-piliated, hyper-adhesive strain of E. coli. We hope that the extreme stickiness of the E. coli will turn them into an excellent bioflocculant- termed EcoGlue. In addition, we will also be expressing a virus protein which specifically binds to the algal species Chlorella vulgaris, a species of interest for harvesting algae oil. This protein will provide additional flocculation ability and specificity for this species. We hope that such a bioflocculant can provide a cheaper, safer alternative to chemical flocculants. |
'''Oil Sands''' | '''Oil Sands''' | ||
| - | Our team will also be working on the [[Oil Sands]] Initiative simultaneously. Tailings ponds have become a major ecological concern regarding oil sands operations due to the toxicity of the water. Oil sands companies are on zero discharge leases, so the tailings water must be held on site, which is done in retention ponds. The main cause of toxicity in tailings water has been identified as naphthenic acids (NAs), some of which can be very recalcitrant to degradation and persist in waters for decades. Thus, our will attempt to expedite the reclamation process. Two bacteria, ''Pseudomonas fluorescens'' and ''Pseudomonas putida'', were isolated from tailings pond sediments and found to be capable of synergistically degrading >95% of a commercial mixture of NAs resembling those found in the tailings water over a 4 week period [ | + | Our team will also be working on the [[Oil Sands]] Initiative simultaneously. Tailings ponds have become a major ecological concern regarding oil sands operations due to the toxicity of the water. Oil sands companies are on zero discharge leases, so the tailings water must be held on site, which is done in retention ponds. The main cause of toxicity in tailings water has been identified as naphthenic acids (NAs), some of which can be very recalcitrant to degradation and persist in waters for decades. Thus, our will attempt to expedite the reclamation process. Two bacteria, ''Pseudomonas fluorescens'' and ''Pseudomonas putida'', were isolated from tailings pond sediments and found to be capable of synergistically degrading >95% of a commercial mixture of NAs resembling those found in the tailings water over a 4 week period [4]. In addition, two other bacteria have shown degradation efficiencies near 100% when in an immobilized cell reactor (ICR)[5], which functionally represents a biofilm. Following these findings, the strategy we wish to implement will be to utilize the two naturally adapted ''Pseudomonas'' species and genetically engineer them to optimize their biofilm formation abilities in tailings pond water-like conditions. This would allow for easier and improved use in an ICR. Alternatively, a more organic and aesthetically pleasing method would be to create a large 'slide' or 'washboard', where the naturally adapted ''Pseudomonas'' species would grow under a constant stream of tailings water, and function similar to an ICR. |
| Line 21: | Line 21: | ||
== Hy-Bi: Pili Hyperexpression == | == Hy-Bi: Pili Hyperexpression == | ||
| - | [[Image:Michigan-Pili2.png|350px|thumb|left|Fig. 1 An electron micrograph view of E. coli, with a close-up of the pili in the inset [ | + | [[Image:Michigan-Pili2.png|350px|thumb|left|Fig. 1 An electron micrograph view of E. coli, with a close-up of the pili in the inset [6].]] |
| - | Type 1 pili (also known as fimbriae) are proteinaceous adhesins that are found on the surface of E. coli. One cell can contain up to 100 pili, which can form up to 2 um long (Fig. 1)[ | + | Type 1 pili (also known as fimbriae) are proteinaceous adhesins that are found on the surface of E. coli. One cell can contain up to 100 pili, which can form up to 2 um long (Fig. 1)[6]. The pili help E. coli form biofilms, and can also be involved in urinary tract infections. However, the strains of E. coli that our team worked with were strictly nonpathogenic. |
| - | The pili are controlled by the ''fim'' operon. This operon consists of several genes, FimA-H. The pili themselves are composed of several thousand subunits of FimA. The tip of each pili consists of the genes FimF, FimG, and FimH. FimH is an adhesin and is linked to FimA through FimF and FimG. Inside the cell, FimC carries proteins to the structural platform, FimD, which then assembles the pilus rod (Fig. 2)[ | + | The pili are controlled by the ''fim'' operon. This operon consists of several genes, FimA-H. The pili themselves are composed of several thousand subunits of FimA. The tip of each pili consists of the genes FimF, FimG, and FimH. FimH is an adhesin and is linked to FimA through FimF and FimG. Inside the cell, FimC carries proteins to the structural platform, FimD, which then assembles the pilus rod (Fig. 2)[7]. This whole process is regulated by the recombinases FimB and FimE. These genes control an invertible DNA sequence, which, when in the "on" position, promotes the production of pili (Fig 3). |
| - | The pili neural network has been characterized in several papers [ | + | The pili neural network has been characterized in several papers [8,9,10]. Essentially, the two recombinases FimB and FimE control an invertible DNA element that acts as a switch, known as FimS. When FimS is in the "on" position, the cell becomes fimbriated. It has been previously determined that the level of piliation depends on the ratio [FimE]/[FimB]. Fig 4 describes the pili regulatory system as a stochastic model. There is only one stable state, when FimB and FimE are both off. After FimB turns on, the cell starts to grow pili and accumulate FimE. When the cell reaches a critical amount of FimE, the cell stops producing pili and the system returns to the stable state. It has been shown that removing the FimE gene will cause the cell to constantly flocculate. |
The reason we are so interested in the pili is their capability to flocculate. Several papers have shown that the pili bind to mannose through FimH. By overproducing the pili, we hope to increase flocculation. We plan to accomplish this goal by putting the FimB gene on a plasmid. You can find the pili team's lab notebook [[Team:Michigan/Pili Expression|here]]. | The reason we are so interested in the pili is their capability to flocculate. Several papers have shown that the pili bind to mannose through FimH. By overproducing the pili, we hope to increase flocculation. We plan to accomplish this goal by putting the FimB gene on a plasmid. You can find the pili team's lab notebook [[Team:Michigan/Pili Expression|here]]. | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | [[Image:Michigan-Pili.png|300px|thumb|left|Fig. 2 A representation of a single pilus and how the different genes in the fim operon function [ | + | [[Image:Michigan-Pili.png|300px|thumb|left|Fig. 2 A representation of a single pilus and how the different genes in the fim operon function [7].]] |
| - | [[Image:Michigan-PiliReg.png|300px|thumb|left|Fig. 3 The recombinases FimB and FimE regulate the fim operon via an invertible segment of DNA [ | + | [[Image:Michigan-PiliReg.png|300px|thumb|left|Fig. 3 The recombinases FimB and FimE regulate the fim operon via an invertible segment of DNA [8].]] |
| - | [[Image:Michigan-PiliReg2.png|200px|thumb|left|Fig. 4 A model that illustrates the different states of the regulatory system [ | + | [[Image:Michigan-PiliReg2.png|200px|thumb|left|Fig. 4 A model that illustrates the different states of the regulatory system [8].]] |
<br style="clear: both" /> | <br style="clear: both" /> | ||
===Results to Date=== | ===Results to Date=== | ||
We were able to put FimB onto pBAD, an arabinose inducible plasmid. This should have resulted in hyperpiliated E. coli, however we were not able to find many pili on our cells when we used an electron microscope. This is discouraging, but it's not a strong indication of failure because our growth conditions may not have been ideal for pili formation. More tests should be done to determine the optimal pili growth conditions. | We were able to put FimB onto pBAD, an arabinose inducible plasmid. This should have resulted in hyperpiliated E. coli, however we were not able to find many pili on our cells when we used an electron microscope. This is discouraging, but it's not a strong indication of failure because our growth conditions may not have been ideal for pili formation. More tests should be done to determine the optimal pili growth conditions. | ||
| - | [[Image: OD_assay_all_samples.png | thumb | 500px|The results of an assay done on [[Team:Michigan/Pili_October#10/19/2010|10/19]]. ]] | + | [[Image: OD_assay_all_samples.png | thumb | 500px|Fig. 5 The results of an assay done on [[Team:Michigan/Pili_October#10/19/2010|10/19]]. ]] |
| - | After conducting several assays to determine the effect of hyperpiliated E. coli on the flocculation of algae, we were not able to see very significant results. Hyperpiliated E. coli seem to flocculate the algae faster than nonpiliated E. coli, but the algae seemed to flocculate by itself. This is possibly due to contaminants in the algae. The algae was not grown in sterile conditions. | + | After conducting several assays to determine the effect of hyperpiliated E. coli on the flocculation of algae, we were not able to see very significant results (Fig. 5). Hyperpiliated E. coli seem to flocculate the algae faster than nonpiliated E. coli, but the algae seemed to flocculate by itself. This is possibly due to contaminants in the algae. The algae was not grown in sterile conditions. |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 55: | Line 55: | ||
The goals of the quorum sensing team are two-fold: first to characterize the response of ''E. coli'' to the ''C. vulgaris'' AI-2 mimic (which may be actual AI-2), and second to engineer ''E. coli'' to flocculate in response to the presence of ''C. vulgaris''. The first task will be accomplished by transforming a LuxS-mutant strain of ''E. coli'' (cannot produce its own AI-2) with an AI-2 reporter biobrick. We will then harvest supernatant from ''C. vulgaris'', which should contain AI-2 or its mimic, and apply it to the reporter strain to test its response. The second task will be accomplished by ligating a gene that causes over-expression of pili (characterized by the pili team) to the Lsr promoter, which is derepressed in response to AI-2. By transforming this part into LuxS-mutant ''E. coli'', we hope to create strain that will stick to algae and will flocculate only in the presence of ''C. vulgaris''. | The goals of the quorum sensing team are two-fold: first to characterize the response of ''E. coli'' to the ''C. vulgaris'' AI-2 mimic (which may be actual AI-2), and second to engineer ''E. coli'' to flocculate in response to the presence of ''C. vulgaris''. The first task will be accomplished by transforming a LuxS-mutant strain of ''E. coli'' (cannot produce its own AI-2) with an AI-2 reporter biobrick. We will then harvest supernatant from ''C. vulgaris'', which should contain AI-2 or its mimic, and apply it to the reporter strain to test its response. The second task will be accomplished by ligating a gene that causes over-expression of pili (characterized by the pili team) to the Lsr promoter, which is derepressed in response to AI-2. By transforming this part into LuxS-mutant ''E. coli'', we hope to create strain that will stick to algae and will flocculate only in the presence of ''C. vulgaris''. | ||
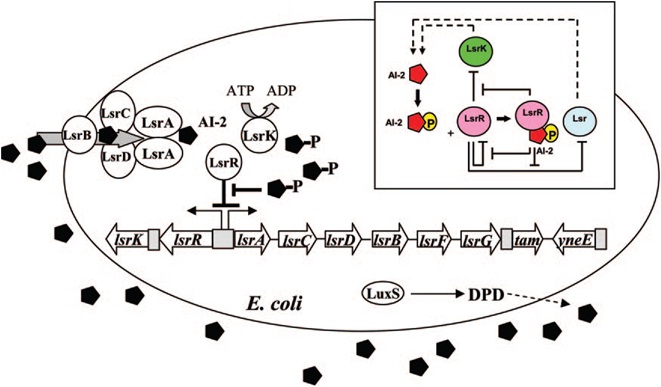
| - | [[Image:QS_circuit.jpg|thumb|700px|left| | + | [[Image:QS_circuit.jpg|thumb|700px|left|Fig. 6 Regulatory mechanisms of the LsrR/phospho-AI-2 circuit in E. coli AI-2 uptake (modified from reference 70). The AI-2 uptake repressor |
LsrR represses many genes including the lsr operon (comprised of lsrACDBFG) and the lsrRK operon. AI-2 can be imported back inside the cell via | LsrR represses many genes including the lsr operon (comprised of lsrACDBFG) and the lsrRK operon. AI-2 can be imported back inside the cell via | ||
LsrACDB. Imported AI-2 is processed as phospho-AI-2 via the kinase LsrK. Phospho-AI-2 has been reported to bind LsrR and relieve its repression | LsrACDB. Imported AI-2 is processed as phospho-AI-2 via the kinase LsrK. Phospho-AI-2 has been reported to bind LsrR and relieve its repression | ||
| - | of the lsr transporter genes, triggering their expression. This in turn stimulates additional AI-2 uptake. DPD, 4,5-dihydroxy-2,3-pentanedione.[ | + | of the lsr transporter genes, triggering their expression. This in turn stimulates additional AI-2 uptake. DPD, 4,5-dihydroxy-2,3-pentanedione.[11]]] |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 85: | Line 85: | ||
== Hy-Bi: Virus Protein Surface Display == | == Hy-Bi: Virus Protein Surface Display == | ||
| - | [[Image:vp130ex.jpg|530px|left|thumb|Fig. | + | [[Image:vp130ex.jpg|530px|left|thumb|Fig.7 The binding of the viral surface protein on algae cells: (A) algae without the virus surface proteins; (B) and (C) algae with virus surface proteins]] |
The idea is to express algae-binding proteins on the surface of E. Coli in order to cause flocculation. Flocculation is the aggregation and precipitation of particles from solution. This flocculation will make the formation of biodiesel more efficient by eliminating the need for centrifugation to concentrate the fuel source. | The idea is to express algae-binding proteins on the surface of E. Coli in order to cause flocculation. Flocculation is the aggregation and precipitation of particles from solution. This flocculation will make the formation of biodiesel more efficient by eliminating the need for centrifugation to concentrate the fuel source. | ||
| - | We devised two ways to cause flocculation, one being using a pili expression and the other using a virus surface protein, and the subject of our group is the virus surface protein. During its viral attack on cells, virus needs a protein that enables it to attach to the surface of the host cells. One of the virus surface proteins, vp 130, is used by chlorovirus to attach itself onto the surface of algae. Onimatsu, et al. recombined the vp 130 gene from Chlorovirus CVK2 with a plasmid, producing a wealth of vp 130. The binding of these proteins on their host cells, chlorella, was detected using fluorescent vp 130 specific antibodies (Fig.6)[ | + | We devised two ways to cause flocculation, one being using a pili expression and the other using a virus surface protein, and the subject of our group is the virus surface protein. During its viral attack on cells, virus needs a protein that enables it to attach to the surface of the host cells. One of the virus surface proteins, vp 130, is used by chlorovirus to attach itself onto the surface of algae. Onimatsu, et al. recombined the vp 130 gene from Chlorovirus CVK2 with a plasmid, producing a wealth of vp 130. The binding of these proteins on their host cells, chlorella, was detected using fluorescent vp 130 specific antibodies (Fig.6)[12]. |
| - | [[Image:vp130.jpg|450px|left|thumb|Fig. | + | [[Image:vp130.jpg|450px|left|thumb|Fig.8 A model of vp 130-expressed bacteria binding algae cells.]] |
So, if we transfer the gene for this surface binding protein from the virus to E. Coli., each bacteria cell will bind to multiple algae cells, which bind to more bacteria, causing aggregation and flocculation. As opposed to using pili as the means to cause flocculation, the advantage of vp130 is its specific binding and aggregation of algae, automatically yielding a high algae mass in the flocculate (Fig.7). However, there are also disadvantages. For example, vp 130 is not a E. Coli surface protein, so it must be cloned as a fusion to a known surface protein. We also suspected OmpA and ice nucleation protein (INP) as candidate surface proteins. | So, if we transfer the gene for this surface binding protein from the virus to E. Coli., each bacteria cell will bind to multiple algae cells, which bind to more bacteria, causing aggregation and flocculation. As opposed to using pili as the means to cause flocculation, the advantage of vp130 is its specific binding and aggregation of algae, automatically yielding a high algae mass in the flocculate (Fig.7). However, there are also disadvantages. For example, vp 130 is not a E. Coli surface protein, so it must be cloned as a fusion to a known surface protein. We also suspected OmpA and ice nucleation protein (INP) as candidate surface proteins. | ||
| Line 100: | Line 100: | ||
== Oil Sands: Ag43 Expression == | == Oil Sands: Ag43 Expression == | ||
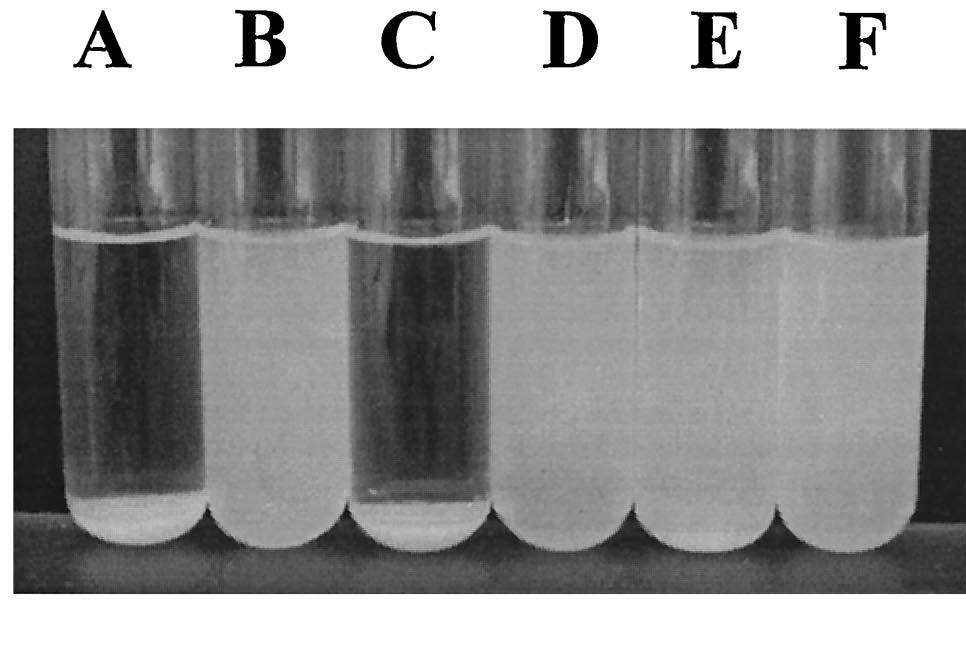
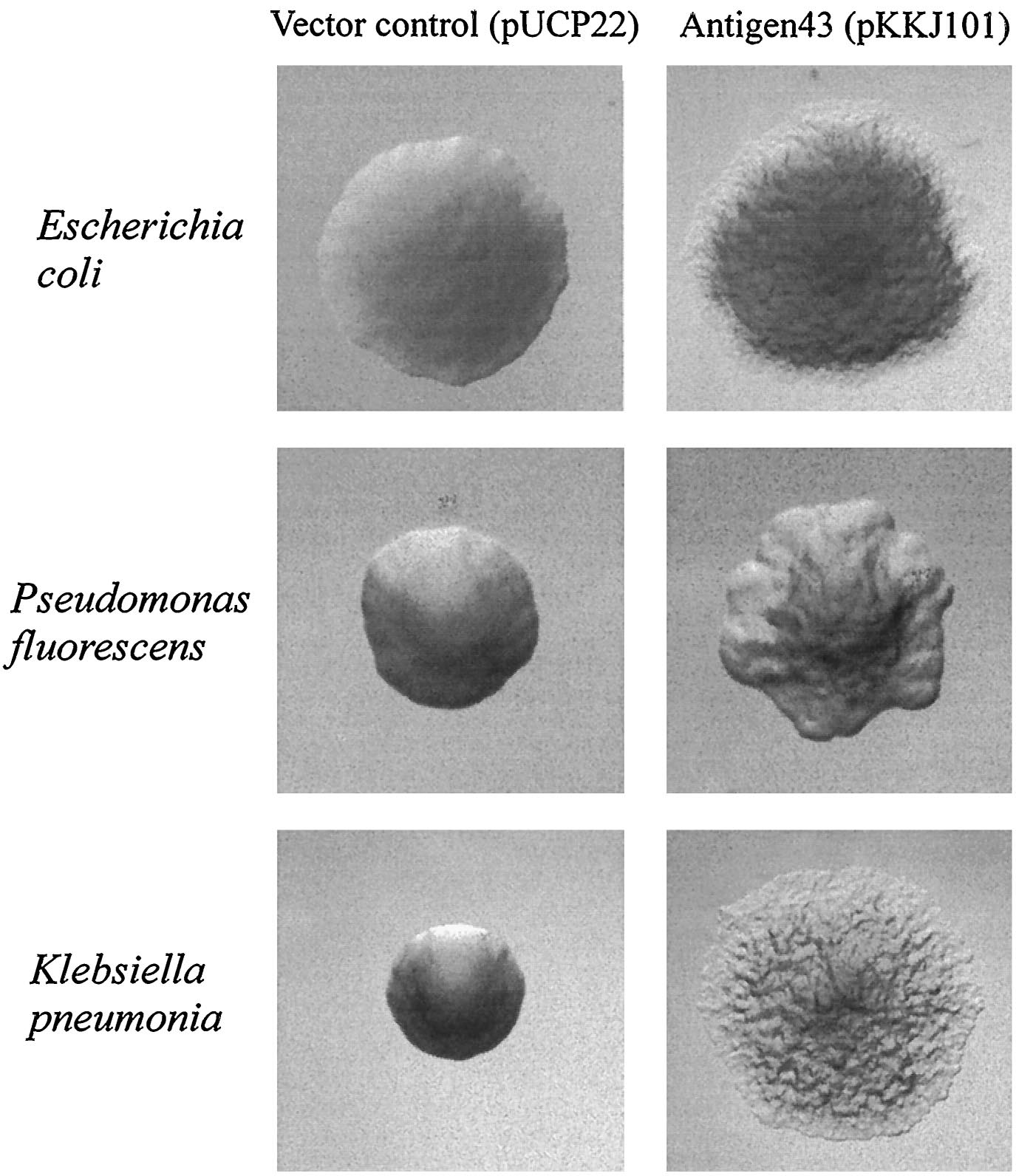
| - | Since the NA degradation pathway in ''Pseudomonas'' is unknown, we decided to improve the biofilm formation abilities of ''Pseudomonas'' based on an early biofilm formation [[Team:Michigan/Oil_Sands_August_September#8/8/2010|crystal violet (CV) assay]]. We decided to clone the ''flu'' gene, which expresses Antigen 43 (Ag43)- a self-dimerizing membrane complex from ''E. coli'', which has been shown to increase the adhesiveness of ''P. fluorescens'' and cause it to form co-species biofilms with ''E. coli'' [ | + | Since the NA degradation pathway in ''Pseudomonas'' is unknown, we decided to improve the biofilm formation abilities of ''Pseudomonas'' based on an early biofilm formation [[Team:Michigan/Oil_Sands_August_September#8/8/2010|crystal violet (CV) assay]]. We decided to clone the ''flu'' gene, which expresses Antigen 43 (Ag43)- a self-dimerizing membrane complex from ''E. coli'', which has been shown to increase the adhesiveness of ''P. fluorescens'' and cause it to form co-species biofilms with ''E. coli'' [13]. The phenotype of ''P. fluorescens'' expressing Ag43 is a "frizzy" colony morphology, increased cell aggregation, and increased adhesiveness to glass surfaces. We hypothesized that this phenotype would improve the ability to use the ''Pseudomonas'' strains (LD1 and LD2) in an immobilized cell reactor (ICR), which has been shown to increase NA degradation 100 fold [5]. |
| - | [[image:Ag43 colonies.gif|thumb|left| | + | [[image:Ag43 colonies.gif|thumb|left|Fig. 9 Colony morphology of cells examined by phase-contrast microscopy. Ag43-expressing strains containing pKKJ101 give rise to changes in colony morphology. |
Strains containing the control plasmid pUCP22 display the same morphotype as the parent strain. Colonies were grown overnight and photographed at the same | Strains containing the control plasmid pUCP22 display the same morphotype as the parent strain. Colonies were grown overnight and photographed at the same | ||
| - | magnification. [ | + | magnification. [13]|400px]] |
| - | [[Image:Ag43 Flocs.png|left|thumb| | + | [[Image:Ag43 Flocs.png|left|thumb|Fig. 10 Settling of cells from |
static liquid suspensions. (A) E. coli HEHA16(pKKJ101), (B) E. coli | static liquid suspensions. (A) E. coli HEHA16(pKKJ101), (B) E. coli | ||
HEHA16(pUCP22), (C) P. fluorescens SBW25(pKKJ101), (D) P. fluorescens | HEHA16(pUCP22), (C) P. fluorescens SBW25(pKKJ101), (D) P. fluorescens | ||
SBW25(pUCP22), (E) K. pneumoniae(pKKJ101), and (F) K. pneumoniae( | SBW25(pUCP22), (E) K. pneumoniae(pKKJ101), and (F) K. pneumoniae( | ||
| - | pUCP22). Note plasmid pKKJ101 expresses the ''flu'' operon. [ | + | pUCP22). Note plasmid pKKJ101 expresses the ''flu'' operon. [13]|400px]] |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 137: | Line 137: | ||
2. http://che.engin.umich.edu/news/savagemakeoil.html | 2. http://che.engin.umich.edu/news/savagemakeoil.html | ||
| - | 3. | + | 3. Lee, A., Lewis, D., Ashman, P. Microbial flocculation, a potentially low-cost harvesting technique for marine microalgae for the production of biodiesel, J Appl Phycol (2009) 21:559–567. |
| - | 4. | + | 4. Del Rio LF, Hadwin a KM, Pinto LJ, MacKinnon MD, Moore MM. Degradation of naphthenic acids by sediment micro-organisms. Journal of applied microbiology. 2006;101(5):1049-61. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17040229. |
| - | 5. | + | 5. Paslawski J, Nemati M, Hill G, Headley J. Biodegradation kinetics of trans -4-methyl-1-cyclohexane carboxylic acid in continuously stirred tank and immobilized cell bioreactors. Journal of Chemical Technology & Biotechnology. 2009;84(7):992-1000. Available at: http://doi.wiley.com/10.1002/jctb.2122. |
| - | 6. | + | 6. Hahn, E., Wild,P., Hermanns, U., Sebbel, P., Glockshuber, R., Haner, M., Taschner, N., Burkhard, P., Aebi, U., A. Muller, S., Exploring the 3D Molecular Architecture of Escherichia coli Type 1 Pili, ''Journal of Molecular Biology'' '''323''' 845-857 (2002) |
| - | 7. | + | 7. Vetsch, M., Puorger, C., Pilus chaperones represent a new type of protein-folding catalyst. ''Nature'' '''431,''' 329-332 (2004). |
| - | 8. | + | 8. Kuwahara, H., Myers, C., Samoilov, M., Abstracted Stochastic Analysis of Type 1 Pili Expression in E. Coli. |
| - | 9. | + | 9. Wolf, D., and Arkin, A., Fifteen Minutes of ''fim'': Control of Type 1 Pili Expression in E. Coli. OMICS 6 2002 |
| - | 10. | + | 10. Schwan, W., Shibata, S., Aizawa, S., and Wolfe, A., The Two-Component Response Regulator RcsB Regulates Type 1 Piliation in Escherichia Coli ''Journal of Bacteriology'' '''189''' 7159-7163 (2007) |
| - | 11. | + | 11. Li, J., Atilla, C., Wang, L., Wood, T. K., Valdes, J. J., Bently, W. E. ''Quorum Sensing in Escherichia coli Is Signaled by AI-2/LsrR: Effects on Small RNA and Biofilm Architecture''. Bacteriology. '''189''': 6011-6020 (2007). |
| - | 12. | + | 12. Onimatsu, H., Sugimoto, I., Fujie, M., Usami, S., Yamada, T. ''Vp 130, a chloroviral surface protein that interacts with the host Chlorella cell wall''. Virology. '''319''': 71-80 (2004). |
13. Kjaergaard K, Schembri M a, Hasman H, Klemm P. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. Journal of bacteriology. 2000;182(17):4789-96. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=111355&tool=pmcentrez&rendertype=abstract. | 13. Kjaergaard K, Schembri M a, Hasman H, Klemm P. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. Journal of bacteriology. 2000;182(17):4789-96. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=111355&tool=pmcentrez&rendertype=abstract. | ||
Latest revision as of 17:04, 27 October 2010
Overall project
Our team worked on two projects simultaneously this year: The Hy-Bi project and the Oil Sands Initiative:
Hy-Bi
The Hy-Bi project is in association with the Chemical Engineering department’s joint project to produce hydrocarbons from algae biomass, dubbed "Hy-Bi"[1,2]. This consists of hydrothermal processing of the algae followed by catalytic upgrading of the “bio crude” oil. Before the hydrothermal processing can take place the algae needs to be concentrated from 1 to 10 g/L to over 250 g/L. Traditional methods that are used like centrifugation, filtration and chemical flocculation are costly.
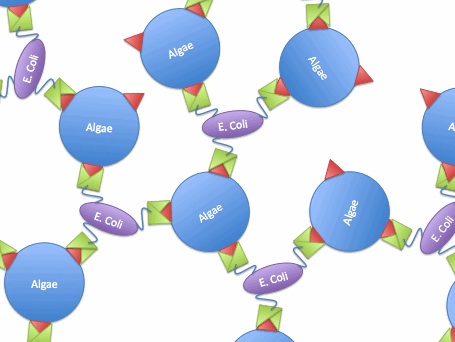
For this project, we examined methods of creating a bioflocculant [3]. We will be over-expressing type I pili and their associated adhesion protein, making a hyper-piliated, hyper-adhesive strain of E. coli. We hope that the extreme stickiness of the E. coli will turn them into an excellent bioflocculant- termed EcoGlue. In addition, we will also be expressing a virus protein which specifically binds to the algal species Chlorella vulgaris, a species of interest for harvesting algae oil. This protein will provide additional flocculation ability and specificity for this species. We hope that such a bioflocculant can provide a cheaper, safer alternative to chemical flocculants.
Oil Sands
Our team will also be working on the Oil Sands Initiative simultaneously. Tailings ponds have become a major ecological concern regarding oil sands operations due to the toxicity of the water. Oil sands companies are on zero discharge leases, so the tailings water must be held on site, which is done in retention ponds. The main cause of toxicity in tailings water has been identified as naphthenic acids (NAs), some of which can be very recalcitrant to degradation and persist in waters for decades. Thus, our will attempt to expedite the reclamation process. Two bacteria, Pseudomonas fluorescens and Pseudomonas putida, were isolated from tailings pond sediments and found to be capable of synergistically degrading >95% of a commercial mixture of NAs resembling those found in the tailings water over a 4 week period [4]. In addition, two other bacteria have shown degradation efficiencies near 100% when in an immobilized cell reactor (ICR)[5], which functionally represents a biofilm. Following these findings, the strategy we wish to implement will be to utilize the two naturally adapted Pseudomonas species and genetically engineer them to optimize their biofilm formation abilities in tailings pond water-like conditions. This would allow for easier and improved use in an ICR. Alternatively, a more organic and aesthetically pleasing method would be to create a large 'slide' or 'washboard', where the naturally adapted Pseudomonas species would grow under a constant stream of tailings water, and function similar to an ICR.
We also decided to split our team into multiple modules that will be working in parallel in order to increase our own efficiency. The work of each module is described in more depth below.
Hy-Bi: Pili Hyperexpression
Type 1 pili (also known as fimbriae) are proteinaceous adhesins that are found on the surface of E. coli. One cell can contain up to 100 pili, which can form up to 2 um long (Fig. 1)[6]. The pili help E. coli form biofilms, and can also be involved in urinary tract infections. However, the strains of E. coli that our team worked with were strictly nonpathogenic.
The pili are controlled by the fim operon. This operon consists of several genes, FimA-H. The pili themselves are composed of several thousand subunits of FimA. The tip of each pili consists of the genes FimF, FimG, and FimH. FimH is an adhesin and is linked to FimA through FimF and FimG. Inside the cell, FimC carries proteins to the structural platform, FimD, which then assembles the pilus rod (Fig. 2)[7]. This whole process is regulated by the recombinases FimB and FimE. These genes control an invertible DNA sequence, which, when in the "on" position, promotes the production of pili (Fig 3).
The pili neural network has been characterized in several papers [8,9,10]. Essentially, the two recombinases FimB and FimE control an invertible DNA element that acts as a switch, known as FimS. When FimS is in the "on" position, the cell becomes fimbriated. It has been previously determined that the level of piliation depends on the ratio [FimE]/[FimB]. Fig 4 describes the pili regulatory system as a stochastic model. There is only one stable state, when FimB and FimE are both off. After FimB turns on, the cell starts to grow pili and accumulate FimE. When the cell reaches a critical amount of FimE, the cell stops producing pili and the system returns to the stable state. It has been shown that removing the FimE gene will cause the cell to constantly flocculate.
The reason we are so interested in the pili is their capability to flocculate. Several papers have shown that the pili bind to mannose through FimH. By overproducing the pili, we hope to increase flocculation. We plan to accomplish this goal by putting the FimB gene on a plasmid. You can find the pili team's lab notebook here.
Results to Date
We were able to put FimB onto pBAD, an arabinose inducible plasmid. This should have resulted in hyperpiliated E. coli, however we were not able to find many pili on our cells when we used an electron microscope. This is discouraging, but it's not a strong indication of failure because our growth conditions may not have been ideal for pili formation. More tests should be done to determine the optimal pili growth conditions.
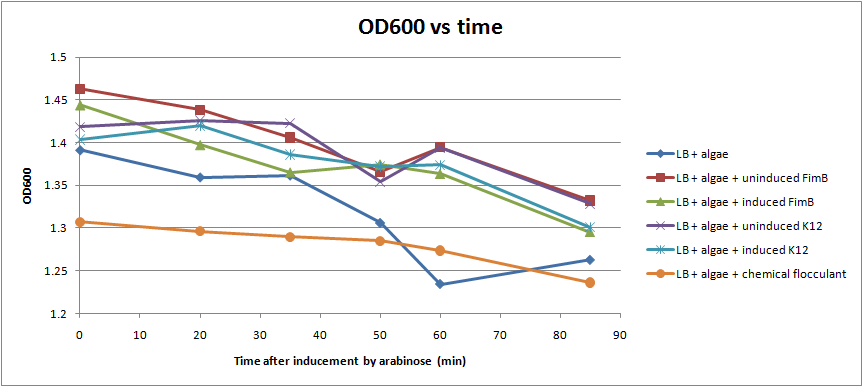
After conducting several assays to determine the effect of hyperpiliated E. coli on the flocculation of algae, we were not able to see very significant results (Fig. 5). Hyperpiliated E. coli seem to flocculate the algae faster than nonpiliated E. coli, but the algae seemed to flocculate by itself. This is possibly due to contaminants in the algae. The algae was not grown in sterile conditions.
Hy-Bi: Quorum Sensing
Our cells will use quorum sensing to determine when the flocculation will start. The cells will produce an inducer either AHL or AI-2. You can find their lab notebook here.
E. coli cells detect their population density by a method known as "quorum sensing." All cells produce a molecule known as Auto-Inducer 2 (AI-2) via the gene LuxS. AI-2 is actively imported via a complex of Lsr membrane proteins, after which it is phosphorylated and binds to the repressor LsrR. This binding relieves the Lsr operon from repression, increasing levels of both the AI-2 import and phosphorylation and of LsrR. The net effect is that cells respond to a threshold concentration of AI-2 (a threshold population density) by altering expression of many genes. E. coli, for example, tends to increase biofilm production in response to AI-2.
Certain species of microalgae useful for harvesting as biofuel, specifically Chlorella vulagris, have been characterized as releasing compounds that mimic the action of AI-2 or AHL, activating the quorum sensing circuits of many bacteria, including Vibrio harveyi, which responds to the same compound as E. coli, AI-2.
The goals of the quorum sensing team are two-fold: first to characterize the response of E. coli to the C. vulgaris AI-2 mimic (which may be actual AI-2), and second to engineer E. coli to flocculate in response to the presence of C. vulgaris. The first task will be accomplished by transforming a LuxS-mutant strain of E. coli (cannot produce its own AI-2) with an AI-2 reporter biobrick. We will then harvest supernatant from C. vulgaris, which should contain AI-2 or its mimic, and apply it to the reporter strain to test its response. The second task will be accomplished by ligating a gene that causes over-expression of pili (characterized by the pili team) to the Lsr promoter, which is derepressed in response to AI-2. By transforming this part into LuxS-mutant E. coli, we hope to create strain that will stick to algae and will flocculate only in the presence of C. vulgaris.
Results to Date
Media:QS_culture_test_8-5-10.pdf
- GFP/YFP activity of AI-2 reporter strains
- MDAI2 is AI-2 knockout - GFP
- W3110 is AI-2 +type - GFP
- DH5α is AI-2 +type - YFP
Media:QS_experiment_8-5-10(1).pdf
- GFP/YFP activity of reporter strains in various supernatants
- column 1 and 2 of plate are negative controls
- column 3 is postive control
- no significant fluorescent activity - we have yet to troubleshoot
- column 4 is experimental: reporter strains in algae culture supernantant
- no significant activity
- repeated above experiment with a stronger AI-2 producing strain as a positive control (BL21)
- No significant results once again
Hy-Bi: Virus Protein Surface Display
The idea is to express algae-binding proteins on the surface of E. Coli in order to cause flocculation. Flocculation is the aggregation and precipitation of particles from solution. This flocculation will make the formation of biodiesel more efficient by eliminating the need for centrifugation to concentrate the fuel source.
We devised two ways to cause flocculation, one being using a pili expression and the other using a virus surface protein, and the subject of our group is the virus surface protein. During its viral attack on cells, virus needs a protein that enables it to attach to the surface of the host cells. One of the virus surface proteins, vp 130, is used by chlorovirus to attach itself onto the surface of algae. Onimatsu, et al. recombined the vp 130 gene from Chlorovirus CVK2 with a plasmid, producing a wealth of vp 130. The binding of these proteins on their host cells, chlorella, was detected using fluorescent vp 130 specific antibodies (Fig.6)[12].
So, if we transfer the gene for this surface binding protein from the virus to E. Coli., each bacteria cell will bind to multiple algae cells, which bind to more bacteria, causing aggregation and flocculation. As opposed to using pili as the means to cause flocculation, the advantage of vp130 is its specific binding and aggregation of algae, automatically yielding a high algae mass in the flocculate (Fig.7). However, there are also disadvantages. For example, vp 130 is not a E. Coli surface protein, so it must be cloned as a fusion to a known surface protein. We also suspected OmpA and ice nucleation protein (INP) as candidate surface proteins.
Results to Date
The cloning for this part was never completed, thus we have no results for this project.
Back to top
Oil Sands: Ag43 Expression
Since the NA degradation pathway in Pseudomonas is unknown, we decided to improve the biofilm formation abilities of Pseudomonas based on an early biofilm formation crystal violet (CV) assay. We decided to clone the flu gene, which expresses Antigen 43 (Ag43)- a self-dimerizing membrane complex from E. coli, which has been shown to increase the adhesiveness of P. fluorescens and cause it to form co-species biofilms with E. coli [13]. The phenotype of P. fluorescens expressing Ag43 is a "frizzy" colony morphology, increased cell aggregation, and increased adhesiveness to glass surfaces. We hypothesized that this phenotype would improve the ability to use the Pseudomonas strains (LD1 and LD2) in an immobilized cell reactor (ICR), which has been shown to increase NA degradation 100 fold [5].
Results to Date
- Relative biofilm formation capacity of different Pseudomonas cultures in different media, with and without napthenic acids at pH7.
- OD readings over 24 hours of the above noted cultures
- Relative biofilm formation capacity of different Pseudomonas cultures in different media, with and without napthenic acids at pH9.
- OD readings over 24 hours of the above noted cultures
Unfortunately, despite having completed some initial control assays, the cloning of Ag43 was never completed, and thus no conclusive results were able to be ascertained.
References
1. http://www.nsf.gov/awardsearch/showAward.do?AwardNumber=0937992
2. http://che.engin.umich.edu/news/savagemakeoil.html
3. Lee, A., Lewis, D., Ashman, P. Microbial flocculation, a potentially low-cost harvesting technique for marine microalgae for the production of biodiesel, J Appl Phycol (2009) 21:559–567.
4. Del Rio LF, Hadwin a KM, Pinto LJ, MacKinnon MD, Moore MM. Degradation of naphthenic acids by sediment micro-organisms. Journal of applied microbiology. 2006;101(5):1049-61. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17040229.
5. Paslawski J, Nemati M, Hill G, Headley J. Biodegradation kinetics of trans -4-methyl-1-cyclohexane carboxylic acid in continuously stirred tank and immobilized cell bioreactors. Journal of Chemical Technology & Biotechnology. 2009;84(7):992-1000. Available at: http://doi.wiley.com/10.1002/jctb.2122.
6. Hahn, E., Wild,P., Hermanns, U., Sebbel, P., Glockshuber, R., Haner, M., Taschner, N., Burkhard, P., Aebi, U., A. Muller, S., Exploring the 3D Molecular Architecture of Escherichia coli Type 1 Pili, Journal of Molecular Biology 323 845-857 (2002)
7. Vetsch, M., Puorger, C., Pilus chaperones represent a new type of protein-folding catalyst. Nature 431, 329-332 (2004).
8. Kuwahara, H., Myers, C., Samoilov, M., Abstracted Stochastic Analysis of Type 1 Pili Expression in E. Coli.
9. Wolf, D., and Arkin, A., Fifteen Minutes of fim: Control of Type 1 Pili Expression in E. Coli. OMICS 6 2002
10. Schwan, W., Shibata, S., Aizawa, S., and Wolfe, A., The Two-Component Response Regulator RcsB Regulates Type 1 Piliation in Escherichia Coli Journal of Bacteriology 189 7159-7163 (2007)
11. Li, J., Atilla, C., Wang, L., Wood, T. K., Valdes, J. J., Bently, W. E. Quorum Sensing in Escherichia coli Is Signaled by AI-2/LsrR: Effects on Small RNA and Biofilm Architecture. Bacteriology. 189: 6011-6020 (2007).
12. Onimatsu, H., Sugimoto, I., Fujie, M., Usami, S., Yamada, T. Vp 130, a chloroviral surface protein that interacts with the host Chlorella cell wall. Virology. 319: 71-80 (2004).
13. Kjaergaard K, Schembri M a, Hasman H, Klemm P. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. Journal of bacteriology. 2000;182(17):4789-96. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=111355&tool=pmcentrez&rendertype=abstract.
 "
"