Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test2settembre
From 2010.igem.org
(→RESULTS (?)) |
m (→RESULTS) |
||
| (3 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
<td valign="top"> | <td valign="top"> | ||
<table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | <table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | ||
| - | <html><p align="center"><font size="5"><b> | + | <html><p align="center"><font size="5"><b>SEPTEMBER, 2ND</b></font></p></html><hr><br> |
==PLATE== | ==PLATE== | ||
<br> | <br> | ||
| Line 43: | Line 43: | ||
* <partinfo>BBa_B0031</partinfo> (negative control) | * <partinfo>BBa_B0031</partinfo> (negative control) | ||
| - | ==RESULTS ( | + | ==RESULTS== |
| + | |||
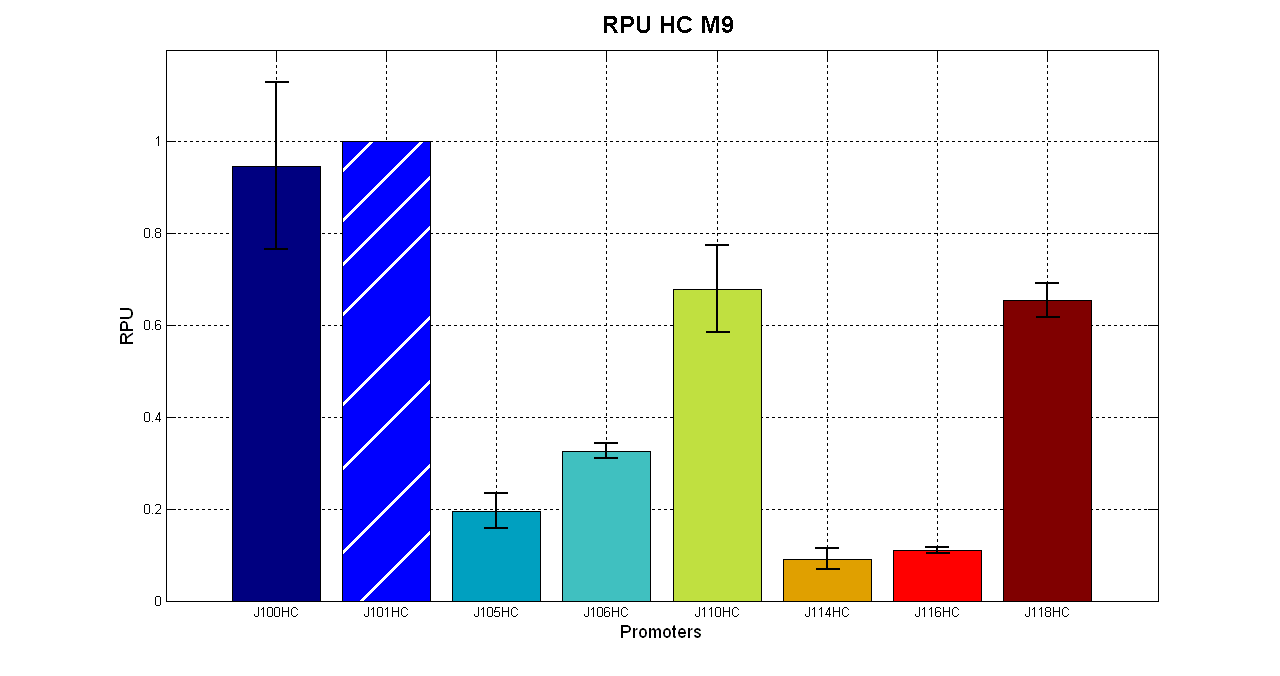
| + | [[Image:UNIPV_Pavia_HCpromotersM9.png| 620px | center ]] | ||
| + | |||
| + | In high copy number plasmids it is possible to identify 3 groups: strong promoters (<partinfo>Ba_J23100</partinfo> and <partinfo>BBa_J23101</partinfo>), medium strength promoters (in order <partinfo>BBa_J23110</partinfo> and <partinfo>BBa_J23118</partinfo>) and a group of weak promoters. | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | || Promoter || R.P.U. (average) || std | ||
| + | |- | ||
| + | || <partinfo>BBa_J23100</partinfo> || 0.946 || 0.182 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23101</partinfo> || 1 || - | ||
| + | |- | ||
| + | || <partinfo>BBa_J23105</partinfo> || 0.196 || 0.038 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23106</partinfo> || 0.326 || 0.015 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23110</partinfo> || 0.678 || 0.096 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23114</partinfo> || 0.091 || 0.022 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23116</partinfo> || 0.101 || 0.004 | ||
| + | |- | ||
| + | || <partinfo>BBa_J23118</partinfo> || 0.655 || 0.035 | ||
| + | |} | ||
| + | |||
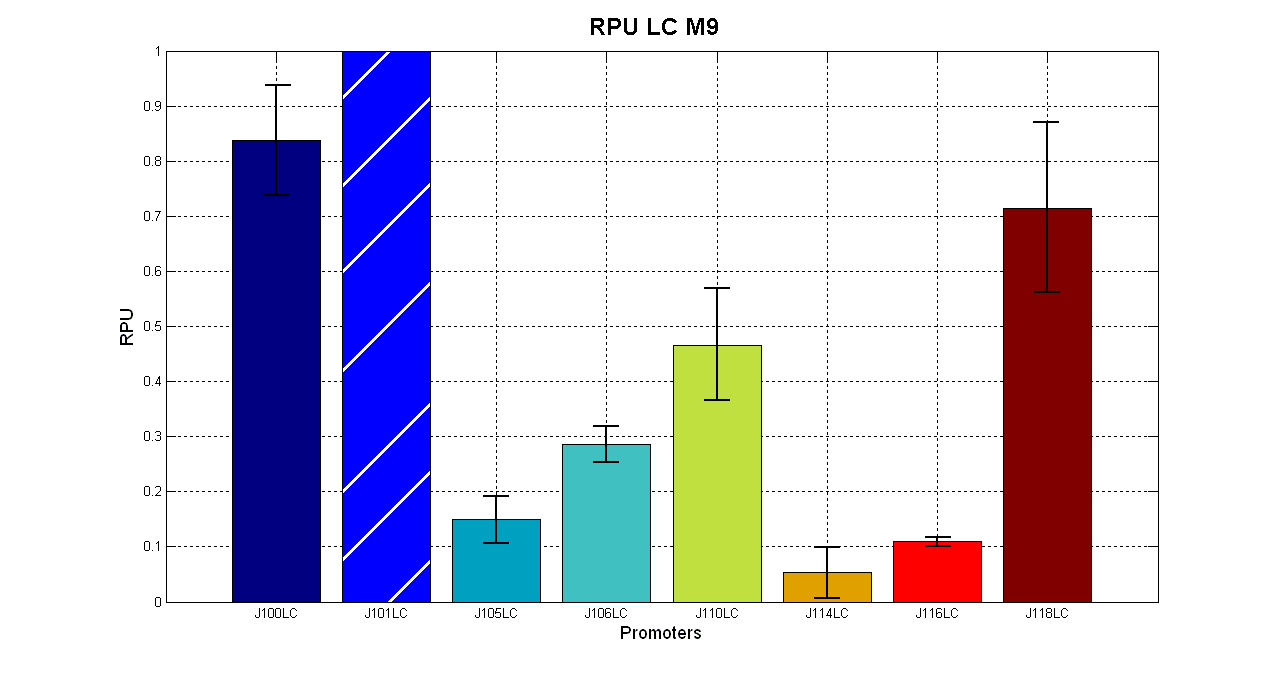
| + | [[Image:UNIPV_Pavia_LCpromotersM9.png| 620px | center ]] | ||
| + | |||
| + | In low copy number plasmids there are the same 3 groups but <partinfo>BBa_J23118</partinfo> resulted stronger than <partinfo>BBa_J23110</partinfo>. | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | || Promoter || R.P.U. (average) || std | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23100</partinfo> || 0.838 || 0.101 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23101</partinfo> || 1 || - | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23105</partinfo> || 0.149 || 0.045 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23106</partinfo> || 0.286 || 0.033 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23110</partinfo> || 0.466 || 0.101 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23114</partinfo> || 0.053 || 0.048 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23116</partinfo> || 0.110 || 0.007 | ||
| + | |- | ||
| + | || <partinfo>pSB4C5</partinfo> - <partinfo>BBa_J23118</partinfo> || 0.714 || 0.154 | ||
| + | |} | ||
<!-- table previous next test --> | <!-- table previous next test --> | ||
Latest revision as of 14:10, 27 October 2010
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"