Team:Michigan/Oil Sands June July
From 2010.igem.org
m (→7/30/2010) |
(→7/30/2010) |
||
| (10 intermediate revisions not shown) | |||
| Line 155: | Line 155: | ||
*Reran gel from 7/10/2010 | *Reran gel from 7/10/2010 | ||
**Lane 1-Invitrogen 1 kb plus ladder | **Lane 1-Invitrogen 1 kb plus ladder | ||
| - | **Lane 2-Digested | + | **Lane 2-Digested BBa_179005 |
| - | ***Ran out of undigested | + | ***Ran out of undigested BBa_179005 |
| - | **Lane 3-Digested | + | **Lane 3-Digested BBa_179015 |
| - | **Lane 4-Uncut miniprep plasmid | + | **Lane 4-Uncut miniprep plasmid BBa_179015 |
We successfully extracted BBa_179015, T7-GFP; expected bands at 906 and 2079 were faintly seen in the gel picture above. No bands appeared for Biobrick Bba_179005, T7 promoter; this biobrick will be transformed again. | We successfully extracted BBa_179015, T7-GFP; expected bands at 906 and 2079 were faintly seen in the gel picture above. No bands appeared for Biobrick Bba_179005, T7 promoter; this biobrick will be transformed again. | ||
| Line 196: | Line 196: | ||
Electroporation Transformation | Electroporation Transformation | ||
| - | All of the transformation plates minus the negative control) grew out! | + | All of the transformation plates (minus the negative control) grew out! |
We should do all transformations by electroporation from now on. Just check with the Lin Lab to make sure we can use the electroporation machine for a few hours before starting. | We should do all transformations by electroporation from now on. Just check with the Lin Lab to make sure we can use the electroporation machine for a few hours before starting. | ||
| Line 208: | Line 208: | ||
***Bba_K103006 | ***Bba_K103006 | ||
***Bba_I719005 | ***Bba_I719005 | ||
| - | **The following biobricks were started from the frozen stock in the - | + | **The following biobricks were started from the frozen stock in the -80°C freezer: |
***BBa_K719015 | ***BBa_K719015 | ||
| Line 222: | Line 222: | ||
''Ann'' | ''Ann'' | ||
| - | An | + | An 8 mL culture of E. coli DH5α in LB broth was produced and placed in the 30°C shaker to grow overnight. |
==7/26/2010== | ==7/26/2010== | ||
| Line 232: | Line 232: | ||
Electroporation transformation | Electroporation transformation | ||
| - | Started larger culture for comp cell preparation in | + | Started larger culture for comp cell preparation in 40 mL of LB in a 500 mL sterile container at 9:00am. |
| - | At 11:30am the | + | At 11:30am the OD600 of the culture was 0.730 and the cultures were placed on ice. |
The transformation of the biobricks was performed according to the electroporation transformation protocol listed under the protocol section of this Wiki. | The transformation of the biobricks was performed according to the electroporation transformation protocol listed under the protocol section of this Wiki. | ||
| Line 267: | Line 267: | ||
The modified miniprep protocol was used listed on the protocol section of the wiki to get a higher DNA concentration. | The modified miniprep protocol was used listed on the protocol section of the wiki to get a higher DNA concentration. | ||
| - | At 9:00pm the GFP, linker and INP parts were miniprepped. The OmpA culture had not grown out (not enough - | + | At 9:00pm the GFP, linker and INP parts were miniprepped. The OmpA culture had not grown out (not enough -80°C freezer stock was added) and a new culture was restarted at 11:00pm in 5 mL of LB with 100 mg/mL AMP. |
'''Pouring Plates''' | '''Pouring Plates''' | ||
| Line 306: | Line 306: | ||
[[Image:7-28biobrickgel.jpg|300px]] | [[Image:7-28biobrickgel.jpg|300px]] | ||
| - | *Lane 1: | + | *Lane 1: Invitrogen 1 kb Plus ladder |
| - | *Lane 2: GFP cut with XbaI and PstI (GFP: 720 bp Backbone: 2079 bp) | + | *Lane 2: GFP cut with XbaI and PstI (GFP: 720 bp; Backbone: 2079 bp) |
*Lane 3: GFP cut with EcoRI and XbaI | *Lane 3: GFP cut with EcoRI and XbaI | ||
*Lane 4: uncut GFP plamid | *Lane 4: uncut GFP plamid | ||
| - | *Lane 5:INP cut with EcoRI and SpeI (INP: 924 bp | + | *Lane 5: INP cut with EcoRI and SpeI (INP: 924 bp; Backbone: 2550 bp) |
| - | *Lane 6:INP cut with SpeI and PstI | + | *Lane 6: INP cut with SpeI and PstI |
| - | *Lane 7:uncut INP plasmid | + | *Lane 7: uncut INP plasmid |
| - | *Lane 8:Linker cut with XbaI and SpeI (Linker: 45 bp | + | *Lane 8: Linker cut with XbaI and SpeI (Linker: 45 bp; Backbone: 2428 bp) |
| - | *Lane 9:Linker cut with SpeI and PstI | + | *Lane 9: Linker cut with SpeI and PstI |
*Lane 10: uncut Linker plasmid | *Lane 10: uncut Linker plasmid | ||
| - | *Lane 11: OmpA cut with EcoRI and SpeI (OmpA: 464 bp | + | *Lane 11: OmpA cut with EcoRI and SpeI (OmpA: 464 bp; Backbone: 2079 bp) |
*Lane 12: OmpA cut with SpeI and PstI | *Lane 12: OmpA cut with SpeI and PstI | ||
| - | *Lane 13:uncut OmpA plasmid | + | *Lane 13: uncut OmpA plasmid |
==7/29/2010== | ==7/29/2010== | ||
| Line 324: | Line 324: | ||
'''Biofilm Assay in LB media''' | '''Biofilm Assay in LB media''' | ||
| - | Cultures of E.coli K12, P. Putida (oilsands) and P. fluorescens (oilsands) were made in 2 mL of LB and placed in the | + | Cultures of E.coli K12, P. Putida (oilsands) and P. fluorescens (oilsands) were made in 2 mL of LB and placed in the 30°C shaker to grow overnight. |
==7/30/2010== | ==7/30/2010== | ||
| Line 330: | Line 330: | ||
'''Biofilm Assay in LB media''' | '''Biofilm Assay in LB media''' | ||
| - | Today the following was carried out: [[Media:7-28-2010_Biofilm_Formation_Experiment.pdf| | + | Today the following was carried out: [[Media:7-28-2010_Biofilm_Formation_Experiment.pdf|Biofilm assay protocol]] |
*@ 8:10am the cultures were started from the overnight | *@ 8:10am the cultures were started from the overnight | ||
| - | *@ 10:00am the OD600 of each culture was measured to be | + | *@ 10:00am the OD600 of each culture was measured to be: |
**E. coli K12: 0.8 | **E. coli K12: 0.8 | ||
**P. fluorescens: 1.2 | **P. fluorescens: 1.2 | ||
| Line 343: | Line 343: | ||
[[Image:7-30-2010_Biofilm_formation_in_LB_media_jpeg.jpg|500px]] | [[Image:7-30-2010_Biofilm_formation_in_LB_media_jpeg.jpg|500px]] | ||
| - | There is a large error in the P. putida strain because there was the largest variation in the | + | There is a large error in the P. putida strain because there was the largest variation in the OD600 of the liquid culture which also resulted in a large variation in the CV OD600 reading. |
We need to find a reference to compare our biofilms to. This could be either a strain that does not form biofilms or a strain that forms biofilms very well. | We need to find a reference to compare our biofilms to. This could be either a strain that does not form biofilms or a strain that forms biofilms very well. | ||
| Line 359: | Line 359: | ||
'''Transformation of pBAD''' | '''Transformation of pBAD''' | ||
| - | The transformation was successful; 30 colonies were observed on the plate. One colony was picked at 7:30 pm for a miniprep the next day and added to 5mL of LB + 25 mg/mL KAN + 1 mM IPTG to grow out in the | + | The transformation was successful; 30 colonies were observed on the plate. One colony was picked at 7:30 pm for a miniprep the next day and added to 5mL of LB + 25 mg/mL KAN + 1 mM IPTG to grow out in the 30°C shaker. |
| - | Another culture was started of GFP from the - | + | Another culture was started of GFP from the -80°C freezer stock in 5 mL of LB + 100 mg/mL AMP for a miniprep. Both the ligations with GFP did not work; as long as we are miniprepping and digesting we want to re-verify this part. |
Latest revision as of 00:19, 27 October 2010
| Sunday | Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | |
| Week 1 | - | 6/28/2010 | 6/29/2010 | 6/30/2010 | 7/1/2010 | 7/2/2010 | - |
| Week 2 | - | - | - | 7/7/2010 | 7/8/2010 | 7/9/2010 | 7/10/2010 |
| Week 3 | - | 7/12/2010 | 7/13/2010 | 7/14/2010 | - | - | - |
| Week 4 | - | - | - | - | - | - | - |
| Week 5 | 7/25/2010 | 7/26/2010 | 7/27/2010 | 7/28/2010 | 7/29/2010 | 7/30/2010 | 7/31/2010 |
Back to Oil Sands Main Notebook
6/28/2010
Pseudomonas putida KT2440 Antibiotic Resistance
- Protocol for starting overnight culture from -80°C freezer
- All protocols can also be found in the Protocols section of the Notebook
6/29/2010
Pseudomonas putida KT2440 Antibiotic Resistance
6/29/2010 and 6/30/2010
Pseudomonas putida KT2440 Antibiotic Resistance
7/1/2010
Literature Review Napthenic Acids Composition & HPLC Analysis
7/2/2010
Pseudomonas putida KT2440 Antibiotic Resistance
7/7/2010
Ann
Biobrick Transformation with Alex and Jennifer
- Made CaCl2 solution and 100 mg/mL amphicilin stock solution according to the media section of the Wiki.
- Started an overnight culture of DH5α according to the heat shock transformation protocol.
- A 12 mL culture was started because the protocol was multiplied by 4.
7/8/2010
Ann
Biobrick Transformation with Marc, Katie and Audra according to the heat shock transformation protocol
- Started culture for Biobrick transformation from overnight at 2:30.
- At 5:00pm the cultures had overgrown to an OD600 of 1.2.
- A 1:3 dilution of cells was performed and the cells were allowed to go through another doubling period of 20 minutes.
- The OD600 was measured again and found to be around 0.500.
- the cultures were placed on ice at 5:30 and allowed to chill for 20 minutes.
- The washings were performed and the comp cells were resuspended in the residual CaCl2 only.
- The OD600 of the comp cells without the glycerol was above 0.3 therefore the comp cells were concentrated enough.
- The heat shock was performed for the following biobricks:
- BBa_179015
- BBa_179005
- BBa_145001
- BBa_117008
- BBa_117002
- BBa_103006
- The cultures were placed in the 30C shaker to grow up for an hour at 30C at 9:00pm.
- The cultures were plated at 10:00pm.
7/9/2010
Ann
Biobrick Transformation with Josh, Prae, Charlie according to the miniprep protocol
- After 9 hours of growing at 37C, the plates did not have any colonies on them.
- After 22 hours of growth plates that had grown had clear colonies.
- The positive and negative controls grew out accordingly.
- Only BBa_179015 and BBa_179005 had colonies (both from plate 1 and none from plate 2...):
- BBa_179015 had over 100 colonies
- BBa_179005 had 2 colonies
- 5 mL overnight cultures were started with 100 mg/mL AMP at 8pm from a single colony on each plate.
7/10/2010
Ann
Biobrick Transformation with Marcus, Bryce, Kilho, Josh, Charlie, Jeremy
Miniprep
- Made frozen stocks from overnight culture of miniprep.
- Performed miniprep on BBa_179015 and BBa_179005.
- Measured DNA concentration with the Nanodrop:
- BBa_179015-11 ng/mL
- BBa_179005-40 ng/mL
Digest
- Ran according to the digest protocol in the protocol section.
- Cut parts with EcoRI and PstI.
- Digested at 37C for 30 minutes.
Gel
- Ran according to the protocol in the protocol section.
- Used 8 well comb instead of 13 well comb; the samples were too dilute too see.
7/12/2010
Ann
Biobrick Transformation with Jeremy
Gel
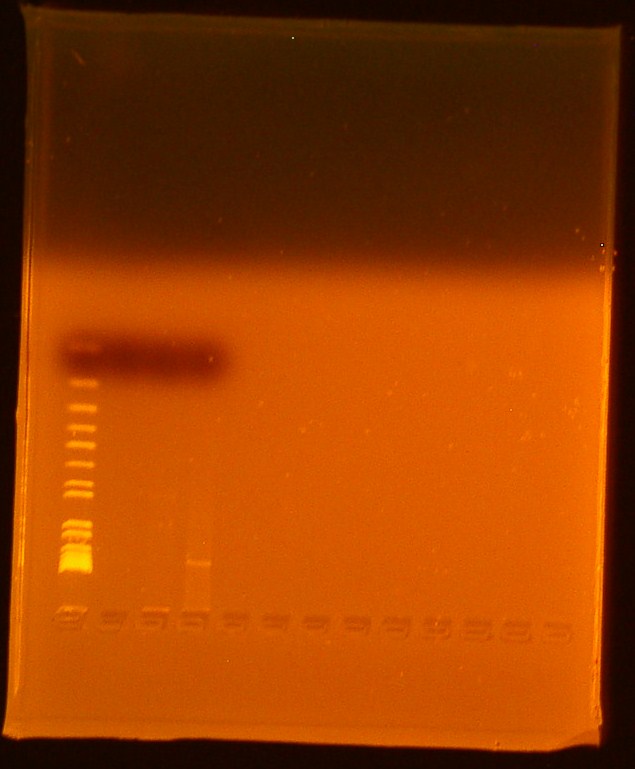
- Reran gel from 7/10/2010
- Lane 1-Invitrogen 1 kb plus ladder
- Lane 2-Digested BBa_179005
- Ran out of undigested BBa_179005
- Lane 3-Digested BBa_179015
- Lane 4-Uncut miniprep plasmid BBa_179015
We successfully extracted BBa_179015, T7-GFP; expected bands at 906 and 2079 were faintly seen in the gel picture above. No bands appeared for Biobrick Bba_179005, T7 promoter; this biobrick will be transformed again.
Biobrick Transformation take 2 with Jeremy
Autoclaved DI water for transformation and autoclaved sterile containers.
- To autoclave sterile containers fill with DI water and decant the water before you start the culture in the container.
7/13/2010
Biobrick Transformation take 2 with Jeremy, Marcus, Josh, Kevin, Audra, Katie
Ann
This was performed according to the electroporation transformation.
- Started the culture at 9:05am
- Removed the culture at 12:05pm with an OD600 of 0.809
- The OD600 of the comp cells were 1.2 after washing
- The time constant for all electroporation were between 5.6 and 5.8 for the following biobricks
- Bba_K117008
- Bba_K117008 #2 (from resuspending remaining part left in registry in 15 uL of ultra pure water)
- Bba_K117002
- Bba_K145001
- Bba_K103006
- Bba_I719005
- The cultures grew in the incubator from 2:15-4:00pm.
- The cultures started to clump after growing this time.
- 100 uL of cells were plated on 100 mg/mL AMP plates.
7/14/2010
Biobrick Transformation take 2
Ann
Electroporation Transformation
All of the transformation plates (minus the negative control) grew out!
We should do all transformations by electroporation from now on. Just check with the Lin Lab to make sure we can use the electroporation machine for a few hours before starting.
Miniprep
- Started overnight cultures in 5 mL of LB plus 100 mg/mL AMP
- The following biobricks were started from the transformation plate:
- Bba_K117008
- Bba_K117002
- Bba_K145001
- Bba_K103006
- Bba_I719005
- The following biobricks were started from the frozen stock in the -80°C freezer:
- BBa_K719015
- The following biobricks were started from the transformation plate:
INPNC Biobrick part
- We received the INPNC Biobrick (Bba_K265008) from UC Davis 2009 team. THANK YOU!!!
- It was shipped on LB plates in a pMA-SK plasmid from Mr. Gene in E. coli DH5α
- Since this plasmid has AMP antibiotic resistance I am pouring plates today with Marc and Kevin using 100 mg/mL AMP resistance; the culture will be streaked out tomorrow with Josh.
7/25/2010
Biobrick Transformation of suface display and pBAD
Ann
An 8 mL culture of E. coli DH5α in LB broth was produced and placed in the 30°C shaker to grow overnight.
7/26/2010
Biobrick Transformation of suface display and pBAD
Ann
Electroporation transformation
Started larger culture for comp cell preparation in 40 mL of LB in a 500 mL sterile container at 9:00am.
At 11:30am the OD600 of the culture was 0.730 and the cultures were placed on ice.
The transformation of the biobricks was performed according to the electroporation transformation protocol listed under the protocol section of this Wiki.
The biobricks removed from the registry are as follows:
- E0040 (GFP)
- K157013 (linker)
- I0500 (pBAD)
The miniprep of I79015 was used as a positive control and a negative control was also run
The OD600 of the comp cells was not measured.
All of the time constants for the transformation were above 5.
The GFP, linker parts and the positive and negative control were plated on 100 mg/mL AMP plates. The pBAD part and negative control were plated on 50 mg/mL KAN plates.
7/27/2010
Biobrick Transformation of surface display and pBAD
Transformation Results
The GFP and linker transformation worked and many colonies grew out on the 100 mg/mL AMP plates.
The pBAD part did not grow out on the KAN plates. Upon further inspection this plasmid needs to be grown with IPTG to switch the origin of replication to a higher copy above 100 copies per cell. Otherwise, another origin of replication dominates that is less than 10 copies per cell. An antibiotic concentration of 50 mg/mL is probably too high; this transformation will have to be attempted a second time.
Miniprep of surface display
At 9:00am a 5 mL culture of the newly transformed GFP part, the newly transformed linker part, the OmpA part transformed previously and the INP from the UC Davis team were started for a miniprep in LB with 100 mg/mL AMP.
The modified miniprep protocol was used listed on the protocol section of the wiki to get a higher DNA concentration.
At 9:00pm the GFP, linker and INP parts were miniprepped. The OmpA culture had not grown out (not enough -80°C freezer stock was added) and a new culture was restarted at 11:00pm in 5 mL of LB with 100 mg/mL AMP.
Pouring Plates
More LB with 100 mg/mL AMP plates and LB with 25 mg/mL KAN plates were poured.
7/28/2010
OmpA Miniprep
The OmpA culture grew out and was miniprepped according the modified miniprep protocol at 11:30am.
Nanodrop of surface display parts
The miniprepped cultures from yesterday were nanodropped to determine the DNA concentration according to the protocol in the protocol section of the wiki:
- GFP: 47.8 ng/uL
- linker: 33.2 ng/uL
- INP: 41.8 ng/uL
- OmpA: 49.3 ng/uL
Digest of surface display parts
The following biobricks were digested according to the protocol on the wiki with the enzymes listed below:
- GFP 1: XbaI and PstI
- GFP 2: EcoRI and XbaI
- INP 1: EcoRI and SpeI
- INP 2: SpeI and PstI
- Linker 1: XbaI and PstI
- Linker 2: SpeI and PstI
- OmpA 1: EcoRI and SpeI
- OmpA 2: SpeI and PstI
Gel of Digest for surface display parts
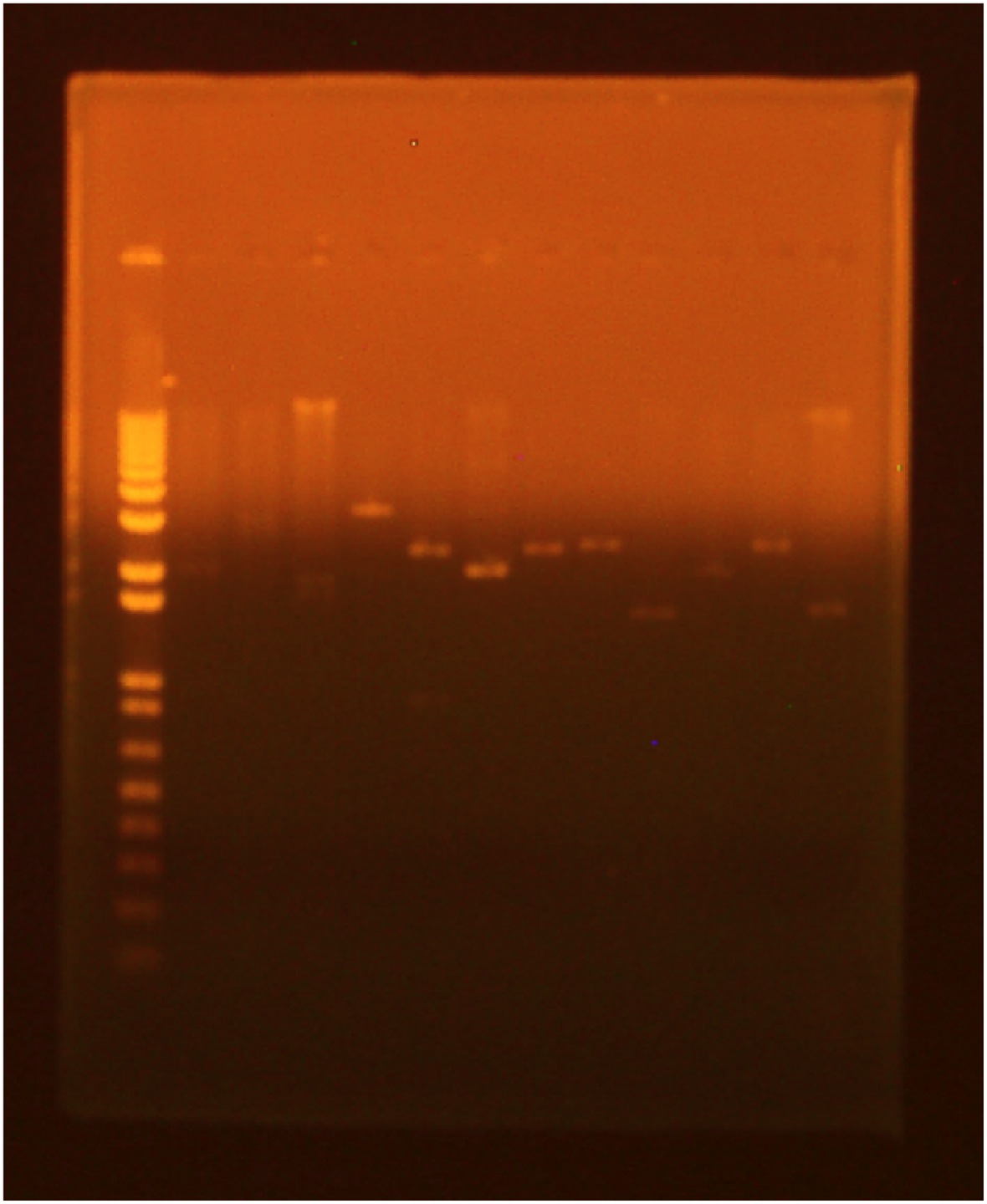
A gel was run of the above digest with uncut plasmid as a control:
- Lane 1: Invitrogen 1 kb Plus ladder
- Lane 2: GFP cut with XbaI and PstI (GFP: 720 bp; Backbone: 2079 bp)
- Lane 3: GFP cut with EcoRI and XbaI
- Lane 4: uncut GFP plamid
- Lane 5: INP cut with EcoRI and SpeI (INP: 924 bp; Backbone: 2550 bp)
- Lane 6: INP cut with SpeI and PstI
- Lane 7: uncut INP plasmid
- Lane 8: Linker cut with XbaI and SpeI (Linker: 45 bp; Backbone: 2428 bp)
- Lane 9: Linker cut with SpeI and PstI
- Lane 10: uncut Linker plasmid
- Lane 11: OmpA cut with EcoRI and SpeI (OmpA: 464 bp; Backbone: 2079 bp)
- Lane 12: OmpA cut with SpeI and PstI
- Lane 13: uncut OmpA plasmid
7/29/2010
Biofilm Assay in LB media
Cultures of E.coli K12, P. Putida (oilsands) and P. fluorescens (oilsands) were made in 2 mL of LB and placed in the 30°C shaker to grow overnight.
7/30/2010
Biofilm Assay in LB media
Today the following was carried out: Biofilm assay protocol
- @ 8:10am the cultures were started from the overnight
- @ 10:00am the OD600 of each culture was measured to be:
- E. coli K12: 0.8
- P. fluorescens: 1.2
- P. putida: 1.0
- These cultures were overgrown but still used for the experiment.
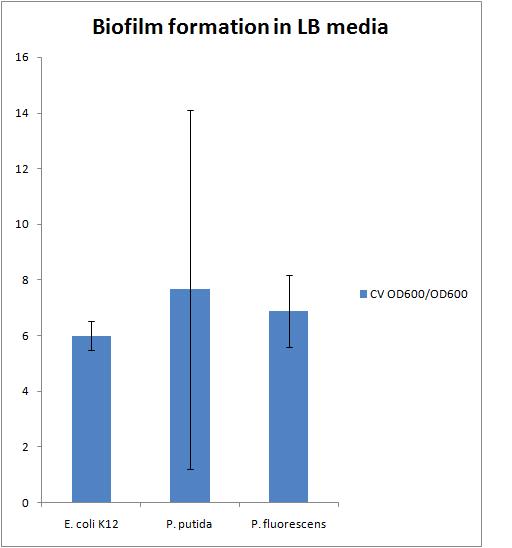
The results of this experiment are presented in the chart below:
There is a large error in the P. putida strain because there was the largest variation in the OD600 of the liquid culture which also resulted in a large variation in the CV OD600 reading.
We need to find a reference to compare our biofilms to. This could be either a strain that does not form biofilms or a strain that forms biofilms very well.
Transformation of pBAD
Marcus and Alex tried the transformation of pBAD again according to the electroporation protocol with Ann's Notes.
The recovery after electroporation was performed in 1 mL of LB with 1mM of IPTG added in order to have the pBAD part have a high copy number.
30 uL of 1M IPTG solution was added to a 25 mg/mL KAN plate and spread with sterile beads in order to keep the high copy plasmid in the pBAD part while plating.
7/30/2010
Transformation of pBAD
The transformation was successful; 30 colonies were observed on the plate. One colony was picked at 7:30 pm for a miniprep the next day and added to 5mL of LB + 25 mg/mL KAN + 1 mM IPTG to grow out in the 30°C shaker.
Another culture was started of GFP from the -80°C freezer stock in 5 mL of LB + 100 mg/mL AMP for a miniprep. Both the ligations with GFP did not work; as long as we are miniprepping and digesting we want to re-verify this part.
 "
"