Team:Michigan/Protocols
From 2010.igem.org
(Difference between revisions)
| (63 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Michigan Header}} | {{Michigan Header}} | ||
| - | |||
| - | + | ==Protocols== | |
| - | + | ||
| - | + | ||
| - | + | '''Using Lab Equipment''' | |
| - | + | ||
| - | [[ | + | [[Media:Protocol_for_Obtaining_Deionized_Water_in_the_ERB.pdf|Obtaining Deionized Water in the ERB]] |
| + | |||
| + | [[Media:ERB_Spectrophotometer_Usage_Protocol.pdf|ERB Spectrophotometer]] | ||
| + | |||
| + | [[Media:7-6-2010_ERB_Autoclave_Liquid_Sterilization_Protocol.pdf|Enigeering Research Building Autoclave]] | ||
| + | |||
| + | [[Media:Epifluorescence_Microscope_Usage_-_Dow.pdf|Epifluorescence Microscope Usage - H.H. Dow]] | ||
| - | |||
'''Cell Culture''' | '''Cell Culture''' | ||
| - | [[Media:6-28-2010_Protocol_for_making_cultures_from_a_-80C_freezer_stock.pdf|Making cultures from a - | + | [[Media:6-28-2010_Protocol_for_making_cultures_from_a_-80C_freezer_stock.pdf|Making cultures from a -80°C freezer stock]] |
[[Media:6-29-2010_Making_Frozen_Stoks.pdf|Making frozen stocks]] | [[Media:6-29-2010_Making_Frozen_Stoks.pdf|Making frozen stocks]] | ||
[[Media:6-27-2010_P._putida_antibiotic_resistance_protocol.pdf|''P. putida'' KT2440 antibotic resistance tolerance]] | [[Media:6-27-2010_P._putida_antibiotic_resistance_protocol.pdf|''P. putida'' KT2440 antibotic resistance tolerance]] | ||
| + | |||
| + | [[Media:Protocol_for_Culturing_Strains_from_CGSC.pdf|Culturing CGSC Strains]] | ||
| + | |||
| + | [[Media:OD600 Cell Dilution Protocol.pdf|OD600 Cell Dilution Protocol]] | ||
| + | |||
| + | [[Media:Quick Transformation of E. coli Competant Cell Preperation.pdf| Competent Cell Preparation for Quick Transformation of E. coli]] | ||
'''DNA Manipulation''' | '''DNA Manipulation''' | ||
| - | [[Media:General_Transformation_Protocol.pdf|Transformation]] | + | [[Media:Direct Plating Transformation Protocol.pdf|Direct Plating Transformation Protocol]] |
| + | *Very simple protocol | ||
| + | *Twice as efficient as standard heat shock | ||
| + | |||
| + | [[Media:Protocol_Heat_Shock_Transformation_2.pdf|Transformation - heat shock]] | ||
| + | |||
| + | *Getting parts from the 360 well registry plates | ||
| + | *Competent cell preparation | ||
| + | *Heat shock | ||
| + | |||
| + | [[Media:General_Transformation_Protocol.pdf|Transformation - electroporation]] | ||
*Getting parts from the 360 well registry plates | *Getting parts from the 360 well registry plates | ||
*Competent cell preparation | *Competent cell preparation | ||
*Electroporation | *Electroporation | ||
| - | [[Media:General_Miniprep_Protocol.pdf|Miniprep]] | + | [[Media:Electroporation Protocol 2.pdf|Electroporation Protocol 2]] |
| + | *Adopted from Jeremy Minty's protocol | ||
| + | *General protocol with specific instructions for ERB/Lin lab equipment | ||
| + | |||
| + | [[Media:PCR Screening Protocol.pdf|PCR Screening Protocol]] | ||
| + | *Efficient protocol for screening transformants after a ligation | ||
| + | |||
| + | [[Media:MODIFIED_Miniprep_Protocol.pdf|MODIFIED Miniprep]] | ||
| + | *[[Media:General_Miniprep_Protocol.pdf|Miniprep]] | ||
| + | |||
| + | [[Media:Nanodrop_Protocol.pdf|Nanodrop]] | ||
[[Media:General_DNA_Digest_Protocol.pdf|DNA Digest]] | [[Media:General_DNA_Digest_Protocol.pdf|DNA Digest]] | ||
| - | [[Media:Protocol_for_Running_a_Gel.pdf| | + | [[Media:_Digest_reagents_calculator.xls|Digest Reagents Calculator]] |
| + | |||
| + | [[Media:Protocol_for_Running_a_Gel.pdf|Gel Electrophoresis]] | ||
[[Media:Protocol_for_DNA_Purification.pdf|DNA Purification]] | [[Media:Protocol_for_DNA_Purification.pdf|DNA Purification]] | ||
| - | [[Media:Ligation_Protocol.pdf|Ligation]] | + | [[Media:Quick_Ligation_Protocol.pdf|Quick Ligation Protocol]] |
| - | * | + | |
| - | *Jeremy Minty Protocol | + | [[Media:8-8-2010_Protocol_for_gel_extraction.pdf|Gel extraction]] |
| + | |||
| + | [[Media:8-10-2010_Ligation_Calculations_updated.pdf|Updated T4 DNA Ligase Protocol (Not quick ligase)]] | ||
| + | *For additional information/questions, refer to [http://www.neb.com/nebecomm/tech_reference/modifying_enzymes/ligation_tips.asp NEB Ligation Tips] | ||
| + | *[[Media:Ligation_Protocol.pdf|Ligation]] | ||
| + | **Ginko Bioworks Protocol | ||
| + | **Jeremy Minty Protocol | ||
| + | ===Group-Specific Protocols=== | ||
| + | ====Oil Sands==== | ||
| + | |||
| + | [[Media:8-6-2010_Biofilm_Formation_Experiment.pdf|Revised biofilm assay protocol]] | ||
| + | *[[Media:7-28-2010_Biofilm_Formation_Experiment.pdf|Biofilm assay protocol]] | ||
| + | |||
| + | [[Media:Static+biofilm+quantification.pdf|Alex's biofilm assay protocol]] | ||
| + | |||
| + | [[Media:7-31-2010_Flu_operon_primers.pdf|General primer design]] | ||
| + | |||
| + | [[Media:7-31-2010_Ag43_flu_gene_WITH_MODIFIED_PRIMER_SITES.pdf|Detailed primer design]] | ||
| + | |||
| + | [[Media:8-4-2010_Colony_PCR_flu.pdf|Colony PCR protocol]] | ||
| + | |||
| + | [[Media:NanR_Cloning.pdf|NanR cloning]] | ||
| + | *Updated Colony PCR Protocol | ||
| + | |||
| + | [[Media:NA Extraction.pdf|NA Extraction Protocol]] | ||
| + | |||
| + | ====Pili==== | ||
| + | [[Media:FimB_Ligation_Protocol.pdf|FimB Ligation]] | ||
| + | |||
| + | [[Media:FimB_PCR.pdf|FimB PCR]] | ||
| + | |||
| + | [[Media:Flocculation_protocol.pdf|Flocculation Protocol]] | ||
| + | |||
| + | ====Quorum Sensing==== | ||
| + | [[Media:Quorum+Sensing+Tables-1-.pdf|Logic Table]] | ||
| + | |||
| + | [[Media:QS_Procedures-1-.pdf|Lsr Circuit Test Protocols]] | ||
| + | |||
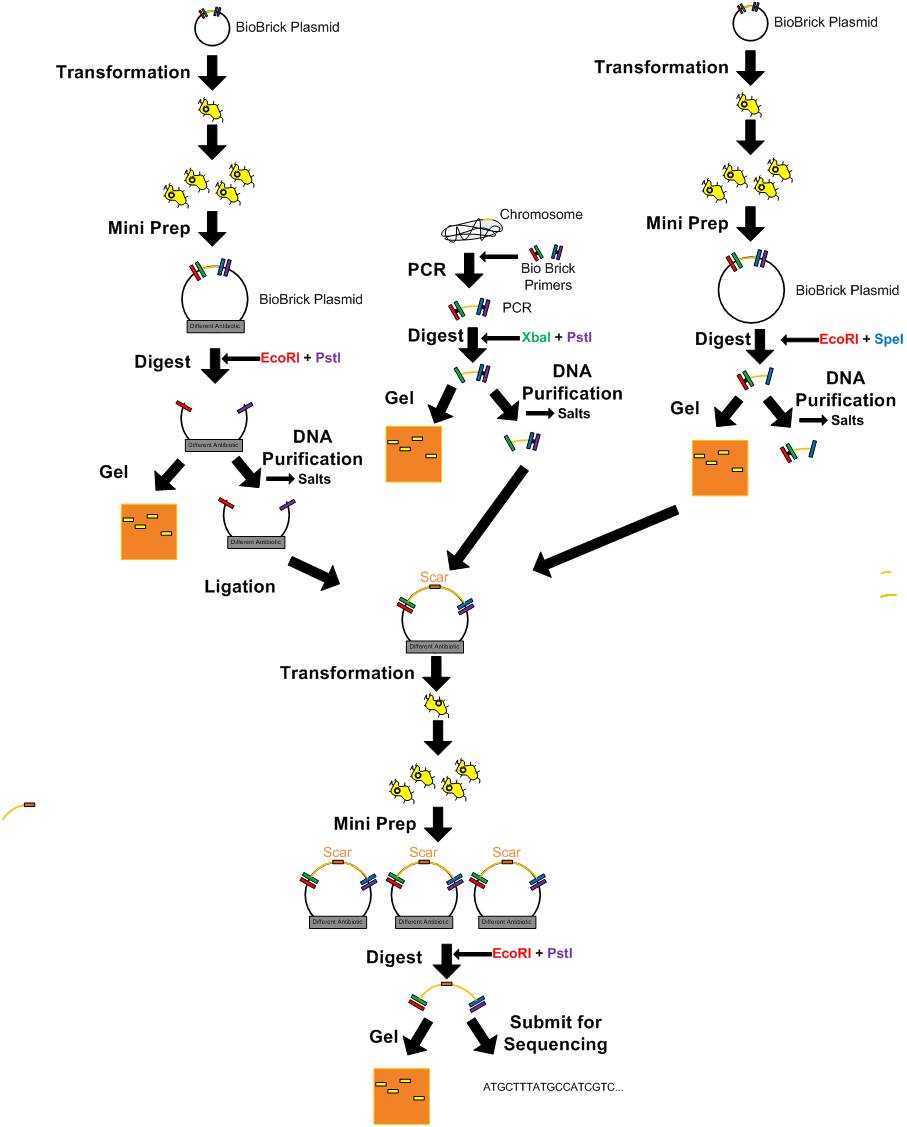
| + | ==General Outline for Biobrick Manipulations== | ||
| + | |||
| + | [[Image:Igem_order_of_procedures.jpg|900px]] | ||
Latest revision as of 00:01, 27 October 2010
Protocols
Using Lab Equipment
Obtaining Deionized Water in the ERB
Enigeering Research Building Autoclave
Epifluorescence Microscope Usage - H.H. Dow
Cell Culture
Making cultures from a -80°C freezer stock
P. putida KT2440 antibotic resistance tolerance
Competent Cell Preparation for Quick Transformation of E. coli
DNA Manipulation
Direct Plating Transformation Protocol
- Very simple protocol
- Twice as efficient as standard heat shock
- Getting parts from the 360 well registry plates
- Competent cell preparation
- Heat shock
Transformation - electroporation
- Getting parts from the 360 well registry plates
- Competent cell preparation
- Electroporation
- Adopted from Jeremy Minty's protocol
- General protocol with specific instructions for ERB/Lin lab equipment
- Efficient protocol for screening transformants after a ligation
Updated T4 DNA Ligase Protocol (Not quick ligase)
- For additional information/questions, refer to [http://www.neb.com/nebecomm/tech_reference/modifying_enzymes/ligation_tips.asp NEB Ligation Tips]
- Ligation
- Ginko Bioworks Protocol
- Jeremy Minty Protocol
Group-Specific Protocols
Oil Sands
Revised biofilm assay protocol
- Updated Colony PCR Protocol
 "
"