Team:Stockholm/27 September 2010
From 2010.igem.org
m |
NinaSchiller (Talk | contribs) (→Concentration measurement) |
||
| Line 142: | Line 142: | ||
===Concentration measurement=== | ===Concentration measurement=== | ||
| + | |||
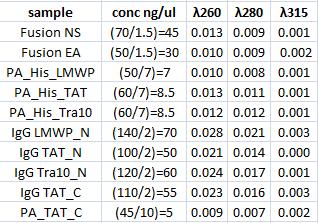
| + | I measured conc of the samples for preparation of the following ligation. | ||
| + | |||
| + | [[Image:Aq31.jpg|300px]] | ||
| + | |||
| + | ===Ligation=== | ||
| + | |||
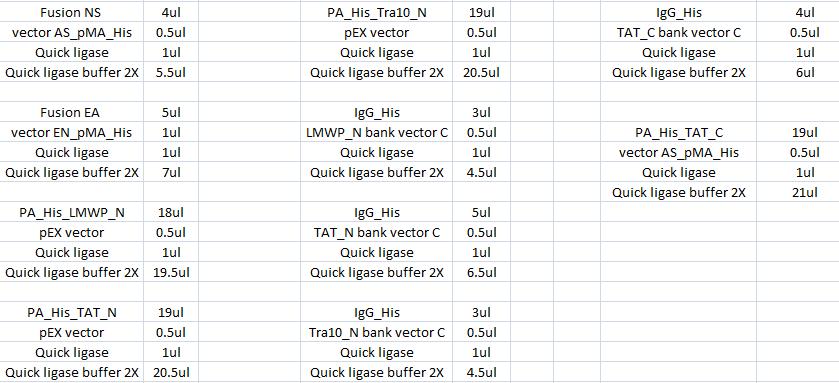
| + | The ligation was according to: | ||
| + | |||
| + | [[Image:Aq32.jpg|700px]] | ||
| + | |||
| + | I incubated the ligations in a water bath with 22 °C in 15 min. | ||
| + | |||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Revision as of 15:25, 26 October 2010
Contents |
Andreas
Cloning and assembly
Transformation results
From 25/9 transformations
Good colony yield on all plates. Good white:red colony ratio on pEX plates (Constructs 1, 2, 3 and 4).
Colony PCR
Picked colonies for colony PCR.
- pEX.N-LMWP⋅SOD⋅His (K): 1-2
- pEX.N-TAT⋅SOD⋅His: 1-2
- pEX.N-Tra10⋅SOD⋅His: 1-2
- pEX.N-LMWP⋅SOD⋅His (C): 1-2
- pSB1K3.N-LMWP⋅SOD⋅His.RBS.yCCS: 1-4
- pSB1K3.N-TAT⋅SOD⋅His.RBS.yCCS: 1-4
- pSB1K3.N-Tra10⋅SOD⋅His.RBS.yCCS: 1-4
- pSB1C3.N-LMWP⋅SOD⋅His.RBS.yCCS: 1-4
- BL21 pEX.N-TAT⋅SOD⋅His 3: 1-2
- BL21 pEX.N-TAT⋅SOD⋅His 4: 1-2
Standard colony PCR protocol.
- Elongation time: 2:00
Gel verification
Gel 1
1 % agarose, 110 V
Gel 2
0.8 % agarose, 90 V
Expected bands
- 744 bp
- 735 bp
- 765 bp
- 744 bp
- 1645 bp
- 1636 bp
- 1666 bp
- 1645 bp
- 735 bp
- 735 bp
Results
Gels run too far (no data). New gels will be run tomorrow.
Sequencing
Samples prepared 25/9 were sent for sequencing. Sequencing information added to the 25/9 notebook page.
Mimmi
Over expression
| Start cultures |
| *3ml LBAMP + tip from glycerol stock |
| *Grow ON in 37°C, 225rpm |
| DNA |
| pEX.SOD |
| pEX.yCCS |
| pEX.SOD.his |
| pEX.his.SOD |
Nina
Sequencing
I send two samples for sequencing. 15 ul sample and 1.5 ul Forward primer.
- pMa #3 Tyrosinase_N ASB0045 694
- Tyrosinase in bank vector K ASB0045 695
Digestion
Digestion of Fusion (1/9):
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme NgoMIV 1 ul
- Restriction enzyme SpeI 1 ul (Add after 1.5 h in 37 °C & incubate in additional 30 min)
Digestion of Fusion (1/9):
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme AgeI 1 ul
- Restriction enzyme EcoRI 1 ul (Add after 1.5 h in 37 °C & incubate in additional 30 min)
Inactivate 20 min 80 °C all samples
Digestion of IgG #5_E_pMa:
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme NgoMIV 1 ul
- Restriction enzyme PstI 1 ul (Add after 1.5 h in 37 °C & incubate in additional 30 min)
Digestion of IgG #5_E_pMa:
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme AgeI 1 ul
- Restriction enzyme EcoRI 1 ul (Add after 1.5 h in 37 °C & incubate in additional 30 min)
Digestion of Protein A#5_E_pMa:
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme AgeI 1 ul
- Restriction enzyme EcoRI 1 ul (Add after 1.5 h in 37 °C & incubate in additional 30 min)
Digestion of Protein A#5_CPPs_N:
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme XbaI 1 ul
- Restriction enzyme PstI 1 ul
Incubate in 37 °C for 30 min.
Concentration measurement
I measured conc of the samples for preparation of the following ligation.
Ligation
The ligation was according to:
I incubated the ligations in a water bath with 22 °C in 15 min.
 |

|
 |

|
 |

|

|

|
 "
"