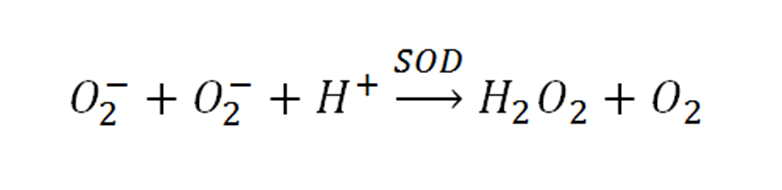Team:Stockholm/8 October 2010
From 2010.igem.org
(New page: {{Stockholm/Top2}} ==Andreas==) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
==Andreas== | ==Andreas== | ||
| + | [[image:Birthdaycake.jpg|Birthday cake]] | ||
| + | |||
| + | '''Short day in lab - birthday celebrations!''' | ||
| + | |||
| + | ===Growth curve assay=== | ||
| + | Inoculated fresh LB with 0.3 mM IPTG with ON cultures to a startup OD of 0.2 and a total volume of 50 ml in 150 ml side-arm E-flasks. | ||
| + | |||
| + | Was advised to use a smaller cultures for the flask size. Experiment canceled after about an hour; will be repeated next week. | ||
| + | |||
| + | ===Removal of insertion in BioBrick suffixes=== | ||
| + | ====Plasmid prep==== | ||
| + | Spun down pSB1C3.SOD ON culture at 13,000 x ''g'', 10 min. Supernatant discarded and cell pellet stored in -20 °C for later plasmid prep. | ||
| + | |||
| + | ==Nina== | ||
| + | |||
| + | ===Protein purification=== | ||
| + | |||
| + | I performed a protein purification of SOD.His (N terminal). The pellet came from an IPTG induced 12 ml overnight culture. | ||
| + | |||
| + | Lysis buffer: | ||
| + | |||
| + | 630 ul * 2 = 1260 ul lysis buffer | ||
| + | |||
| + | Wash buffer: | ||
| + | |||
| + | 10X lysis buffer 12.6 ml | ||
| + | |||
| + | Elution buffer: | ||
| + | |||
| + | 5X lysis buffer 6.3 ml | ||
| + | |||
| + | *PMSF | ||
| + | |||
| + | 100 * volume = 1260 * 1 | ||
| + | |||
| + | 12.6 ul PMSF added in lysis buffer | ||
| + | |||
| + | *Imidazole | ||
| + | |||
| + | 2 * volume = 1260 * 10*10^-3 | ||
| + | |||
| + | 6.3 ul imidazole added in lysis buffer | ||
| + | |||
| + | *Imidazole | ||
| + | |||
| + | 2 * volume = 1260 * 20*10^-3 | ||
| + | |||
| + | 126 ul imidazole added in wash buffer | ||
| + | |||
| + | *Lysozyme | ||
| + | |||
| + | The tip of a spoon was added in lysis buffer | ||
| + | |||
| + | *DNase | ||
| + | |||
| + | 20 ug/ml was added in lysis buffer | ||
| + | |||
| + | ====column equilibration==== | ||
| + | |||
| + | The work is carried out in a cold room. | ||
| + | |||
| + | *Ni-resin was vortexed and 1 ml was added in a drop column. | ||
| + | *5 ml wash buffer was added and run through the column (to equilibrate the Ni-resin). | ||
| + | *The lysis mixed with the bacteria pellet was added in the column. However, the mixture seemed to plug the column and nothing could run through. | ||
| + | |||
| + | I left this in the cold room over the weekend to check with anyone at the department what to do and if I still can work with this material. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === SOD activity assay === | ||
| + | |||
| + | *Searching "web of science" for SOD activity assays | ||
| + | |||
| + | [[Image:SOD_activity.png|left|]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | *Generates O<sub>2</sub><sup>-</sup> | ||
| + | **Xanthine + xanthine oxidase | ||
| + | **NADH + phenazine methosulphate | ||
| + | **riboflavin + methionine | ||
| + | |||
| + | |||
| + | *Reacts with O<sub>2</sub><sup>-</sup> and can be measured | ||
| + | **cytochrome C | ||
| + | **nitro-blue tetrazodium NBT | ||
| + | **adrenaline | ||
| + | **pyrogallol | ||
| + | **hydroxylamine | ||
| + | |||
| + | |||
| + | |||
| + | *Catalase-like test? | ||
| + | **No, since E. coli are catalase positive and SOD differences will not be detectable | ||
| + | |||
| + | |||
| + | |||
| + | *SOD activity test that chemically produces O<sub>2</sub><sup>-</sup> | ||
| + | |||
| + | {| | ||
| + | ! mix | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | graded levels of SOD | ||
| + | | ? | ||
| + | | rowspan="9" | | ||
| + | *Incubate in RT, darkness for 10min | ||
| + | |||
| + | *Measure A<sub>540</sub> | ||
| + | |||
| + | *negative control <sup>= without NADH</sup> | ||
| + | |||
| + | |- | ||
| + | | NBT | ||
| + | | 1mg | ||
| + | |- | ||
| + | | phenazine methosulphate | ||
| + | | 10µg | ||
| + | |- | ||
| + | | EDTA | ||
| + | | 1µM | ||
| + | |- | ||
| + | | gelatin | ||
| + | | 1mg | ||
| + | |- | ||
| + | | phosphate | ||
| + | | 0.1M | ||
| + | |- | ||
| + | | NADH 1mM | ||
| + | | 0.1ml | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 3ml | ||
| + | |- | ||
| + | | pH=7.8 | ||
| + | |} | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 11:08, 26 October 2010
Contents |
Andreas
Short day in lab - birthday celebrations!
Growth curve assay
Inoculated fresh LB with 0.3 mM IPTG with ON cultures to a startup OD of 0.2 and a total volume of 50 ml in 150 ml side-arm E-flasks.
Was advised to use a smaller cultures for the flask size. Experiment canceled after about an hour; will be repeated next week.
Removal of insertion in BioBrick suffixes
Plasmid prep
Spun down pSB1C3.SOD ON culture at 13,000 x g, 10 min. Supernatant discarded and cell pellet stored in -20 °C for later plasmid prep.
Nina
Protein purification
I performed a protein purification of SOD.His (N terminal). The pellet came from an IPTG induced 12 ml overnight culture.
Lysis buffer:
630 ul * 2 = 1260 ul lysis buffer
Wash buffer:
10X lysis buffer 12.6 ml
Elution buffer:
5X lysis buffer 6.3 ml
- PMSF
100 * volume = 1260 * 1
12.6 ul PMSF added in lysis buffer
- Imidazole
2 * volume = 1260 * 10*10^-3
6.3 ul imidazole added in lysis buffer
- Imidazole
2 * volume = 1260 * 20*10^-3
126 ul imidazole added in wash buffer
- Lysozyme
The tip of a spoon was added in lysis buffer
- DNase
20 ug/ml was added in lysis buffer
column equilibration
The work is carried out in a cold room.
- Ni-resin was vortexed and 1 ml was added in a drop column.
- 5 ml wash buffer was added and run through the column (to equilibrate the Ni-resin).
- The lysis mixed with the bacteria pellet was added in the column. However, the mixture seemed to plug the column and nothing could run through.
I left this in the cold room over the weekend to check with anyone at the department what to do and if I still can work with this material.
Mimmi
SOD activity assay
- Searching "web of science" for SOD activity assays
- Generates O2-
- Xanthine + xanthine oxidase
- NADH + phenazine methosulphate
- riboflavin + methionine
- Reacts with O2- and can be measured
- cytochrome C
- nitro-blue tetrazodium NBT
- adrenaline
- pyrogallol
- hydroxylamine
- Catalase-like test?
- No, since E. coli are catalase positive and SOD differences will not be detectable
- SOD activity test that chemically produces O2-
| mix | ||
|---|---|---|
| graded levels of SOD | ? |
|
| NBT | 1mg | |
| phenazine methosulphate | 10µg | |
| EDTA | 1µM | |
| gelatin | 1mg | |
| phosphate | 0.1M | |
| NADH 1mM | 0.1ml | |
| tot | 3ml | |
| pH=7.8 |
 |

|
 |

|
 |

|

|

|
 "
"