Team:Stockholm/14 September 2010
From 2010.igem.org
(Difference between revisions)
(→Andreas) |
m |
||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
==Andreas== | ==Andreas== | ||
===Preparation of Top10 chemically competent cells=== | ===Preparation of Top10 chemically competent cells=== | ||
| Line 134: | Line 135: | ||
====Ligations==== | ====Ligations==== | ||
| - | [Dig RBS 34 14/9] = 50 ng/μl | + | *[Dig RBS 34 14/9] = 50 ng/μl |
| - | [Dig pSB1K3 14/9] = 66.6 ng/μl | + | *[Dig pSB1K3 14/9] = 66.6 ng/μl |
| - | [Dig SOD⋅His 14/9] = 66.6 ng/μl | + | *[Dig SOD⋅His 14/9] = 66.6 ng/μl |
| - | [Dig His⋅SOD 14/9] = 66.6 ng/μl | + | *[Dig His⋅SOD 14/9] = 66.6 ng/μl |
| - | [Dig N-TAT 14/9] = 66.6 ng/μl | + | *[Dig N-TAT 14/9] = 66.6 ng/μl |
| - | [Dig N-Tra10 14/9] = 66.6 ng/μl | + | *[Dig N-Tra10 14/9] = 66.6 ng/μl |
| - | [Dig SOD 14/9] = 15 ng/μl | + | *[Dig SOD 14/9] = 15 ng/μl |
{|border="1" cellpadding="1" cellspacing="0" | {|border="1" cellpadding="1" cellspacing="0" | ||
| Line 199: | Line 200: | ||
*15 min on ice | *15 min on ice | ||
*50 μl IPTG (on pEX.SOD Amp 100 plate only) | *50 μl IPTG (on pEX.SOD Amp 100 plate only) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === MITF-M === | ||
| + | |||
| + | ==== FB PCR ==== | ||
| + | |||
| + | {| | ||
| + | ! mix | ||
| + | | (µl)x2 | ||
| + | | rowspan="10" width="100" | | ||
| + | | | ||
| + | | rowspan="10" width="100" | | ||
| + | | colspan="2" | | ||
| + | | rowspan="4" | | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 22.5 | ||
| + | ! primers | ||
| + | ! colspan="2" | Conditions | ||
| + | |- | ||
| + | | F primer | ||
| + | | 1 | ||
| + | | MITF_FB_F_18aug | ||
| + | ! time | ||
| + | ! °C | ||
| + | |- | ||
| + | | R primer | ||
| + | | 1 | ||
| + | | MITF_FB_R_18aug | ||
| + | | 10m | ||
| + | | 95 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 0.5 | ||
| + | | rowspan="6" | | ||
| + | | 30s | ||
| + | | 95 | ||
| + | | ) | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 25µl | ||
| + | | 30s | ||
| + | | 55 | ||
| + | | > 5 cycles | ||
| + | |- | ||
| + | | colspan="2" rowspan="4" | | ||
| + | | 1m40s | ||
| + | | 72 | ||
| + | | ) | ||
| + | |- | ||
| + | | 30s | ||
| + | | 95 | ||
| + | | \ | ||
| + | |- | ||
| + | | 2m10s | ||
| + | | 72 | ||
| + | | / 25 cycles | ||
| + | |- | ||
| + | | 10m | ||
| + | | 72 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ==== Digestion ==== | ||
| + | |||
| + | :::- MITF-M and pSB1C3 | ||
| + | |||
| + | {| | ||
| + | ! Mix | ||
| + | ! MITF | ||
| + | ! pSB1C3 cut S | ||
| + | ! pSB1C3 cut E | ||
| + | | rowspan="7" width="100" | | ||
| + | ! colspan="2" | conditions | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 5 | ||
| + | | ) | ||
| + | | ) | ||
| + | ! time | ||
| + | ! °C | ||
| + | |- | ||
| + | | DNA | ||
| + | | 20 | ||
| + | | >24 | ||
| + | | >24 | ||
| + | | 30m | ||
| + | | 37 | ||
| + | |- | ||
| + | | 10xbuffer | ||
| + | | 3 | ||
| + | | ) | ||
| + | | ) | ||
| + | | 20m | ||
| + | | 65 | ||
| + | |- | ||
| + | | EcoRI | ||
| + | | 1 | ||
| + | | 1 | ||
| + | | | ||
| + | | rowspan="3" | | ||
| + | | rowspan="3" | | ||
| + | |- | ||
| + | | SpeI | ||
| + | | 1 | ||
| + | | | ||
| + | | 1 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 30µl | ||
| + | | 25µl | ||
| + | | 25µl | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==== Gel ==== | ||
| + | |||
| + | [[Image:2010-09-14_MITF_nu_jaklar.jpg|200px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | MITF-M | ||
| + | |- | ||
| + | | 3 | ||
| + | | MITF-M | ||
| + | |- | ||
| + | | 4 | ||
| + | | MITF-M cut E+S | ||
| + | |- | ||
| + | | 5 | ||
| + | | pSB1C3 cut E+S | ||
| + | |- | ||
| + | | 6 | ||
| + | | pSB1C3 cut E+S | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === SOD.his / his.SOD / yCCS === | ||
| + | |||
| + | *preparing for glycerol stock and over-expression... | ||
| + | |||
| + | {| border="1" | ||
| + | | SOD.his | ||
| + | | x2 | ||
| + | | 3ml | ||
| + | |- | ||
| + | | his.SOD | ||
| + | | x2 | ||
| + | | 3ml | ||
| + | |- | ||
| + | | yCCS | ||
| + | | x2 | ||
| + | | 3ml | ||
| + | |} | ||
| + | |||
| + | *Grow at 37°C ON (~175rpm) | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 11:03, 26 October 2010
Contents |
Andreas
Preparation of Top10 chemically competent cells
No growth on neither Amp 100, Cm 25 nor Km 50 plates - no contamination of competent cells.
pSB1C3.N-TAT & .N-Tra10
Successful transformation. One colony of each plate picked and dissolved in 10 μl and saved in fridge for later glycerol stock preparation.
Assembly of new parts
Planned and designed cloning strategies for some of the assemblies I will be constructing:
- pEX.SOD
- N-TAT⋅SOD⋅His
- N-Tra10⋅SOD⋅His
- N-LMWP⋅SOD⋅His (N-LMWP n/a)
- His⋅SOD⋅C-Tra10 (C-Tra10 n/a)
- His⋅SOD⋅C-TAT (C-TAT n/a)
- His⋅SOD⋅C-LMWP (C-LMWP n/a)
- RBS.yCCS
Digestions
- [pSB1C3.SOD] = 105.5 ng/μl
- [pSB1A2.RBS 34] = 60 ng/μl
- [pMA.His⋅SOD] = 266 ng/μl
- [pMA.SOD⋅His] = 246 ng/μl
- [pSB1C3.N-TAT] = 200 ng/μl
- [pSB1C3.N-Tra10] = 151 ng/μl
- [pSB1K3.RFP] = 104 ng/μl
| His⋅SOD | SOD⋅His | RBS 34 | pSB1K3 | N-TAT | N-Tra10 | SOD | |
|---|---|---|---|---|---|---|---|
| 10X FD buffer | 3 | 3 | 3 | 3 | 3 | 3 | 1 |
| dH2O | 16.9 | 17.5 | 0 | 5.8 | 15 | 11.8 | 7 |
| DNA | 8.1 | 7.5 | 25 | 19.2 | 10 | 13.2 | 1 |
| NgoMIV (NEB) | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| PstI | 1 | 0 | 1 | 1 | 0 | 0 | 1 |
| FD SpeI | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| FD EcoRI | 0 | 1 | 0 | 1 | 1 | 1 | 0 |
| AgeI | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| FD XbaI | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 30 μl | 30 μl | 30 μl | 30 μl | 30 μl | 30 μl | 10 μl |
- Incubation: 37 °C
- 2:00 (conventional enzymes)
- 0:30 (FastDigest enzymes)
- Inactivation: 80 °C, 20 min
Ligations
- [Dig RBS 34 14/9] = 50 ng/μl
- [Dig pSB1K3 14/9] = 66.6 ng/μl
- [Dig SOD⋅His 14/9] = 66.6 ng/μl
- [Dig His⋅SOD 14/9] = 66.6 ng/μl
- [Dig N-TAT 14/9] = 66.6 ng/μl
- [Dig N-Tra10 14/9] = 66.6 ng/μl
- [Dig SOD 14/9] = 15 ng/μl
| Lig pEX.SOD 14/9 | Lig pK.N-TAT⋅ SOD⋅His 14/9 | Lig pK.N-Tra10⋅ SOD⋅His 14/9 | Lig pA.RBS. yCCS 14/9 | |
|---|---|---|---|---|
| 10X T4 ligase buffer | 2 | 2 | 2 | 2 |
| Vector DNA | 3 | 1.5 | 1.5 | 2 |
| Insert DNA (1) | 10 | 11 | 11 | 6 |
| Insert DNA (2) | – | 4.5 | 4.5 | – |
| dH2O | 4 | 0 | 0 | 9 |
| T4 DNA ligase | 1 | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl | 20 μl |
- Incubation: 22 °C, 15 min
Transformation
Standard transformation
- 1 μl ligation mix
- 15 min on ice
- 50 μl IPTG (on pEX.SOD Amp 100 plate only)
Mimmi
MITF-M
FB PCR
| mix | (µl)x2 | ||||||
|---|---|---|---|---|---|---|---|
| sH2O | 22.5 | primers | Conditions | ||||
| F primer | 1 | MITF_FB_F_18aug | time | °C | |||
| R primer | 1 | MITF_FB_R_18aug | 10m | 95 | |||
| DNA | 0.5 | 30s | 95 | ) | |||
| tot | 25µl | 30s | 55 | > 5 cycles | |||
| 1m40s | 72 | ) | |||||
| 30s | 95 | \ | |||||
| 2m10s | 72 | / 25 cycles | |||||
| 10m | 72 | ||||||
Digestion
- - MITF-M and pSB1C3
| Mix | MITF | pSB1C3 cut S | pSB1C3 cut E | conditions | ||
|---|---|---|---|---|---|---|
| sH2O | 5 | ) | ) | time | °C | |
| DNA | 20 | >24 | >24 | 30m | 37 | |
| 10xbuffer | 3 | ) | ) | 20m | 65 | |
| EcoRI | 1 | 1 | ||||
| SpeI | 1 | 1 | ||||
| tot | 30µl | 25µl | 25µl | |||
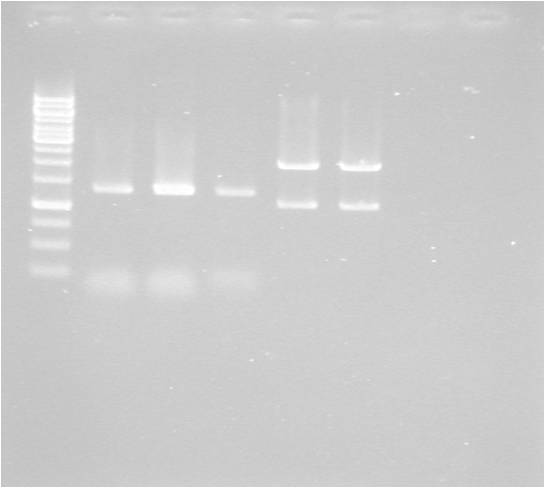
Gel
| well | sample |
|---|---|
| 1 | ladder |
| 2 | MITF-M |
| 3 | MITF-M |
| 4 | MITF-M cut E+S |
| 5 | pSB1C3 cut E+S |
| 6 | pSB1C3 cut E+S |
SOD.his / his.SOD / yCCS
- preparing for glycerol stock and over-expression...
| SOD.his | x2 | 3ml |
| his.SOD | x2 | 3ml |
| yCCS | x2 | 3ml |
- Grow at 37°C ON (~175rpm)
 |

|
 |

|
 |

|

|

|
 "
"