Team:Stockholm/29 July 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Sequencing of SOD) |
m |
||
| (9 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
| - | A streak of colonies from the IgG plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). ~10 colonies from the bFGF plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). This was incubated with shake in 37 °C until OD reaches around 0.6. | + | The transformation from yesterday had gone well. A streak of colonies from the IgG plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). ~10 colonies from the bFGF plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). This was incubated with shake in 37 °C until OD reaches around 0.6. |
[[Image:OD-igg.jpg]] | [[Image:OD-igg.jpg]] | ||
| + | -S = without sonication | ||
| + | +S = with sonication | ||
| + | |||
| + | IPTG treatment at 0, 1, 2 and 3 hours after the first step: | ||
| + | |||
| + | 1. Add 1M IPTG (1ul IPTG/ml bacterial sample) at OD 0.4-0.6 | ||
| + | |||
| + | 2. Pellet 1 ml of the sample | ||
| + | |||
| + | 3. Remove the LB | ||
| + | |||
| + | 4. Resuspend with 50 ul H20 and add 50 ul RDSB | ||
| + | |||
| + | 5. Leave in freezer until loading the samples in a gel for running a coomassie | ||
| + | |||
| + | RDSB: | ||
| + | |||
| + | 100 ul 1M DTT in 900 ul werner's sample buffer | ||
---- | ---- | ||
| + | |||
===PCR on Tyrosinase=== | ===PCR on Tyrosinase=== | ||
| Line 28: | Line 47: | ||
2 = 3 min | 2 = 3 min | ||
| + | |||
| + | PCR reaction mix: | ||
| + | |||
| + | * 1 µl Morten's polymerase PjuX7 | ||
| + | |||
| + | * 1 µl 10 mM dNTPs | ||
| + | |||
| + | * 3 µl 5 µM forward primer | ||
| + | |||
| + | * 3 µl 5 µM revers primer | ||
| + | |||
| + | * 10 µl buffer 5X | ||
| + | |||
| + | * 1 µl MgCl2 50mM | ||
| + | |||
| + | * 30 µl H2O | ||
| + | |||
| + | * DNA template was one colony | ||
| + | |||
| + | PCR program: | ||
| + | |||
| + | 98°C - 2 min | ||
| + | |||
| + | 31 cycles of: | ||
| + | |||
| + | * 98°C - 10 sec | ||
| + | |||
| + | * 55°C - 15 sec | ||
| + | |||
| + | * 72°C - 2,5 min and 3 min | ||
| + | |||
| + | 72°C - 5 min | ||
| + | |||
| + | 4°C - ∞ | ||
| + | |||
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas | DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas | ||
| Line 36: | Line 90: | ||
---- | ---- | ||
| + | |||
===Sequencing of SOD=== | ===Sequencing of SOD=== | ||
I prepared tubes of SOD for sequencing: | I prepared tubes of SOD for sequencing: | ||
| - | 15 ul plasmid from a mini-prep and 1.5 ul (10uM) primer. Preferably the forward primer ( | + | 15 ul plasmid from a mini-prep and 1.5 ul (10uM) primer. Preferably the forward primer (VF2) of the vectors verification primers. |
* SOD A: ASB0045 179 | * SOD A: ASB0045 179 | ||
| Line 46: | Line 101: | ||
---- | ---- | ||
| + | |||
===Colony PCR of CPP=== | ===Colony PCR of CPP=== | ||
| - | I made a colony PCR of CPP (Transportan 10) in two versions. One with the reverse primer of the CPP (Tral_CN_R1) and the vector's forward verification primer (VF2), and the other by combining both of the vector's verification primers (VF2 and VR). | + | The transformation from yesterday had gone well. I made a colony PCR of CPP (Transportan 10) in two versions. One with the reverse primer of the CPP (Tral_CN_R1) and the vector's forward verification primer (VF2), and the other by combining both of the vector's verification primers (VF2 and VR). |
The stock of the reverse primer of CPP (Tral_CN_R1) was diluted 10 times. 105uM to 10 uM by mixing 3 ul of the stock with 27 ul of H2O. This was calculated for 5 CPP colonies on a dish. | The stock of the reverse primer of CPP (Tral_CN_R1) was diluted 10 times. 105uM to 10 uM by mixing 3 ul of the stock with 27 ul of H2O. This was calculated for 5 CPP colonies on a dish. | ||
| Line 55: | Line 111: | ||
---- | ---- | ||
| + | |||
===Mini-prep of bFGF=== | ===Mini-prep of bFGF=== | ||
| Line 62: | Line 119: | ||
The method was according to the procedure described under Protocols. | The method was according to the procedure described under Protocols. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === Site mutated PCR === | ||
| + | |||
| + | {| border="1" cellspacing="0" | ||
| + | ! Mix | ||
| + | | (µl) | ||
| + | | X8 (µl) | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 22.5 | ||
| + | | 180 | ||
| + | |- | ||
| + | | F primer | ||
| + | | 1 | ||
| + | | 8 | ||
| + | |- | ||
| + | | R primer | ||
| + | | 1 | ||
| + | | 8 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 0.5 | ||
| + | | 8X0.5 | ||
| + | |- | ||
| + | | align="right" | '''tot''' | ||
| + | | 25 | ||
| + | | 200 | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| | ||
| + | ! Primer | ||
| + | | width="150" | | ||
| + | ! colspan="2" | conditions | ||
| + | | width="150" | | ||
| + | ! colspan="2" | samples | ||
| + | |- | ||
| + | | pSB1_VF2 | ||
| + | | | ||
| + | ! time | ||
| + | ! °C | ||
| + | | | ||
| + | | c1 | ||
| + | | *yCCSB 1 | ||
| + | |- | ||
| + | | pSB1_VR | ||
| + | | | ||
| + | | 2min | ||
| + | | 94 | ||
| + | | | ||
| + | | c2 | ||
| + | | *yCCSB 2 | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 30s | ||
| + | | 94 | ||
| + | | ) | ||
| + | | A | ||
| + | | *yCCS • A | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 30s | ||
| + | | 60 | ||
| + | | > 30 cycles | ||
| + | | B | ||
| + | | *yCCS • B | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 2m | ||
| + | | 72 | ||
| + | | ) | ||
| + | | C | ||
| + | | *yCCS • C | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | 10m | ||
| + | | 72 | ||
| + | | | ||
| + | | D | ||
| + | | *yCCS • D | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | oo | ||
| + | | 10 | ||
| + | | | ||
| + | | E | ||
| + | | *yCCS • E | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | c | ||
| + | | *blank control | ||
| + | |} | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:56, 26 October 2010
Contents |
Andreas
CPP troubleshooting
Analyzed sequencing results of our constructed CPPs (LMWP and Transportan 10). It seems like our target vector recircularizes instead of ligating with our CPP primers. Me, Johan and Nina are working on troubleshooting this, however it looks like we will soon have to synthesize our genes, since our primer ligations just don't seem to work.
Nina
Protein expression of IgG protease from pEX
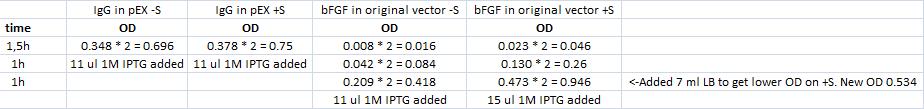
BL21 cultures of IgG in pEX and bFGF in original vector from plates used to inoculate for IPTG induction:
The transformation from yesterday had gone well. A streak of colonies from the IgG plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). ~10 colonies from the bFGF plate was inoculated in two separate falcon tubes containing 12 ml LB and 24 ul ampicillin (50 mg/ml). This was incubated with shake in 37 °C until OD reaches around 0.6.
-S = without sonication
+S = with sonication
IPTG treatment at 0, 1, 2 and 3 hours after the first step:
1. Add 1M IPTG (1ul IPTG/ml bacterial sample) at OD 0.4-0.6
2. Pellet 1 ml of the sample
3. Remove the LB
4. Resuspend with 50 ul H20 and add 50 ul RDSB
5. Leave in freezer until loading the samples in a gel for running a coomassie
RDSB:
100 ul 1M DTT in 900 ul werner's sample buffer
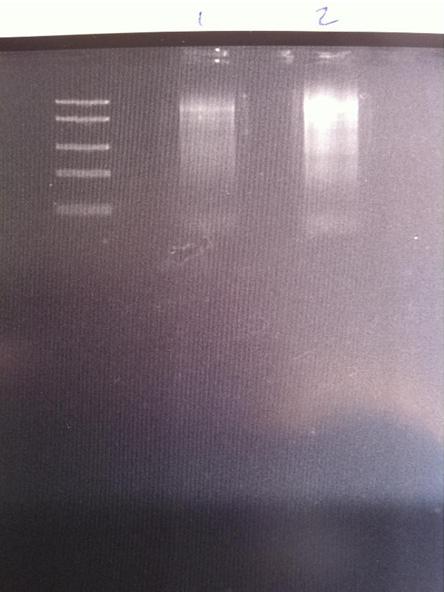
PCR on Tyrosinase
I have had trouble obtaining a band representing tyrosinase on a 1 % agarose gel after running a PCR. Therefore I ran two new PCR:s, one with an annealing time of 2.5 min and another with 3 min.
1 = 2.5 min
2 = 3 min
PCR reaction mix:
- 1 µl Morten's polymerase PjuX7
- 1 µl 10 mM dNTPs
- 3 µl 5 µM forward primer
- 3 µl 5 µM revers primer
- 10 µl buffer 5X
- 1 µl MgCl2 50mM
- 30 µl H2O
- DNA template was one colony
PCR program:
98°C - 2 min
31 cycles of:
- 98°C - 10 sec
- 55°C - 15 sec
- 72°C - 2,5 min and 3 min
72°C - 5 min
4°C - ∞
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas
It looks like I got a correct band on both samples, however the 2.5 min looks better than the other. Unfortunately I also have a lot of unspecific binding.
Sequencing of SOD
I prepared tubes of SOD for sequencing:
15 ul plasmid from a mini-prep and 1.5 ul (10uM) primer. Preferably the forward primer (VF2) of the vectors verification primers.
- SOD A: ASB0045 179
- SOD B: ASB0045 180
Colony PCR of CPP
The transformation from yesterday had gone well. I made a colony PCR of CPP (Transportan 10) in two versions. One with the reverse primer of the CPP (Tral_CN_R1) and the vector's forward verification primer (VF2), and the other by combining both of the vector's verification primers (VF2 and VR).
The stock of the reverse primer of CPP (Tral_CN_R1) was diluted 10 times. 105uM to 10 uM by mixing 3 ul of the stock with 27 ul of H2O. This was calculated for 5 CPP colonies on a dish.
The PCR ran 35 cycles with an annealing time of 30 seconds since the CPP:s are small in size.
Mini-prep of bFGF
Five colonies of bFGF ligated into the shipping vector (with chloramphenicol resistans 50 mg/ml) had been picked by Johan and inoculated in LB ON. I carried out a mini-prep on these five samples.
Samples with colony nr: 1, 3, 4, 5 and 7.
The method was according to the procedure described under Protocols.
Mimmi
Site mutated PCR
| Mix | (µl) | X8 (µl) |
|---|---|---|
| H2O | 22.5 | 180 |
| F primer | 1 | 8 |
| R primer | 1 | 8 |
| DNA | 0.5 | 8X0.5 |
| tot | 25 | 200 |
| Primer | conditions | samples | ||||
|---|---|---|---|---|---|---|
| pSB1_VF2 | time | °C | c1 | *yCCSB 1 | ||
| pSB1_VR | 2min | 94 | c2 | *yCCSB 2 | ||
| 30s | 94 | ) | A | *yCCS • A | ||
| 30s | 60 | > 30 cycles | B | *yCCS • B | ||
| 2m | 72 | ) | C | *yCCS • C | ||
| 10m | 72 | D | *yCCS • D | |||
| oo | 10 | E | *yCCS • E | |||
| c | *blank control | |||||
 |

|
 |

|
 |

|

|

|
 "
"