Team:Heidelberg/Modeling
From 2010.igem.org
AlejandroHD (Talk | contribs) |
AlejandroHD (Talk | contribs) |
||
| Line 5: | Line 5: | ||
==shRNA binding sites== | ==shRNA binding sites== | ||
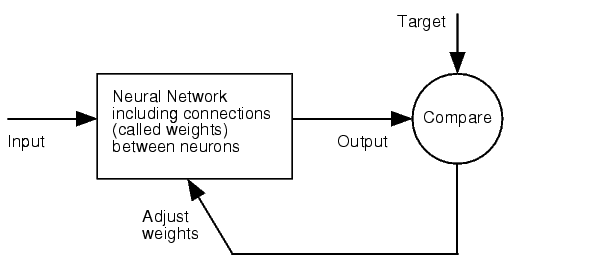
As the title of our project states, “DNA is not enough”. There are several upper-level regulation systems in superior organisms. Our main idea was using miRNA to tune down the expression of genes, having tissue-specific, exactly tuned gene therapy as objective. | As the title of our project states, “DNA is not enough”. There are several upper-level regulation systems in superior organisms. Our main idea was using miRNA to tune down the expression of genes, having tissue-specific, exactly tuned gene therapy as objective. | ||
| + | |||
miRNA are non-coding regulatory RNAs functioning as post-transcriptional gene silencers. After they are processed, they are usually 22 nucleotides long and they usually bind to the 3’UTR region of the mRNA (although they can also bind to the ORF or to the 5'UTR), forcing the mRNA into degradation or just repressing translation. | miRNA are non-coding regulatory RNAs functioning as post-transcriptional gene silencers. After they are processed, they are usually 22 nucleotides long and they usually bind to the 3’UTR region of the mRNA (although they can also bind to the ORF or to the 5'UTR), forcing the mRNA into degradation or just repressing translation. | ||
| + | |||
In vegetal organisms, miRNA usually bind to the mRNA with extensive complementarity. In animals, interactions are more inexact, creating a lot of uncertainty in the in silico prediction of targets. | In vegetal organisms, miRNA usually bind to the mRNA with extensive complementarity. In animals, interactions are more inexact, creating a lot of uncertainty in the in silico prediction of targets. | ||
| + | |||
| + | The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA, and it usually requires extensive pairing. (For the sake of simplicity, we extended slightly the term seed to include the nucleotides 1-8.) | ||
| + | |||
| + | Outside the seed, the existence of supplemental pairing (at least 3 contiguous nucleotides and centered in nucleotides 13-16 of the miRNA) stabilizes the bound complex and increases the efficacy of the binding site. | ||
| + | |||
| + | Binding sites with a high local AU density around the binding site have proven to be more effective (possibly because of the destabilization of the mRNA secondary structure around the site). | ||
Targetscan Scores Jan ???? | Targetscan Scores Jan ???? | ||
| Line 16: | Line 24: | ||
Apart from these two options, the user can personalize the binding site to meet their individual requirements. | Apart from these two options, the user can personalize the binding site to meet their individual requirements. | ||
| - | ===Seed types=== | + | ====Seed types==== |
| - | + | In miBS designer, the user can choose between several types of seed for their binding site (ordered by increasing efficacy): | |
| - | + | ||
| + | - 6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. | ||
| + | |||
| + | - 7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. | ||
| + | |||
| + | - 7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. | ||
| + | |||
| + | - 8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. | ||
| + | |||
| + | - Apart from any of these options, the user can decide to create a customized seed with a mismatch included. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
*The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | *The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | ||
| - | ===Supplementary region=== | + | ====Supplementary region==== |
| - | + | In miBS designer, the user can choose among several types of supplementary sequences, starting with 3 matching nucleotides (14-16), increasing sequentially until 8 (13-20), and then total matching (from 13-22, leaving a bulge). In case the user needs some other specific supplementary region, he can customize the sequence by inputting the desired matching nucleotides. | |
| - | + | ||
| - | ===AU content=== | + | ====AU content==== |
| - | + | In order to allow the user to improve the efficacy of their binding sites, miBS designer offers options to increase the AU content by adding adenine or uracil to positions around the matches (specifically in -1, 0, 1, 8, 9 and 10). The function is designed so that it varies the AU content without introducing new pairings. | |
| - | ===Sticky | + | ====Sticky ends==== |
| - | In order to facilitate the task of introducing the binding site into a plasmid, the user can add sequences to both | + | In order to facilitate the task of introducing the binding site into a plasmid, the user can add sequences to both ends of the binding site. Initially, the user can choose among the RFC-12 standard for biobricks BB2, the XmaI/XhoI restriction enzymes used somewhere in the project, or some custom sequences input by the user. In the last case, the output sequences will not be directly ready for cloning: the user has to either digest the construction prior to ligation, or to process the primers before ordering them to remove the extra nucleotides. |
===Output=== | ===Output=== | ||
Revision as of 05:46, 26 October 2010

|
|
||||||||||||
 "
"