Team:Heidelberg/Modeling
From 2010.igem.org
(→miRockdown on miBEAT) |
(→miRockdown on miBEAT) |
||
| Line 55: | Line 55: | ||
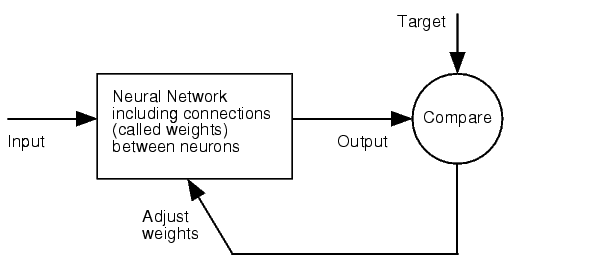
Right from the beginning of our modeling project, we knew we would have to integrate our trained models into an online GUI. We realized it in the most user friendly way we could think of: The user only needs to input the desired knockdown percentage (kd%) and choose an sh/miRNA sequence, to get a binding site that satisfies the users needs.<br> | Right from the beginning of our modeling project, we knew we would have to integrate our trained models into an online GUI. We realized it in the most user friendly way we could think of: The user only needs to input the desired knockdown percentage (kd%) and choose an sh/miRNA sequence, to get a binding site that satisfies the users needs.<br> | ||
<br> | <br> | ||
| - | [[Image:MiRockdownOverview.png|thumb| | + | [[Image:MiRockdownOverview.png|thumb|600px|left|Overview of the miRockdown script flow: The knockdown percentage (kd%) input invokes the selection of the right experimental and model binding site or binding site parameters respectively. The binding site (BS) sequence input starts the generation of on the fly generated BS sequences, which are characterized by a modified targetscan_scores algorithm. The parameters of the selected model BS are correlated with the generated BS parameters and the most similar of the generated BS is the output.]] |
<br> | <br> | ||
The results of both of our models and the experimentally verified binding sites are integrated in [miRockdown] (see Figure: miRockdown) on the [miBEAT] GUI. For every binding site request of a user there are the results of the three different concepts displayed. Thus the users can always choose which of the three differently generated binding to use. The binding site with the most similar experimentally observed knockdown percentage is given out, together with its properties and oligos ready to clone into the [https://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit miTuner]-construct.<br> | The results of both of our models and the experimentally verified binding sites are integrated in [miRockdown] (see Figure: miRockdown) on the [miBEAT] GUI. For every binding site request of a user there are the results of the three different concepts displayed. Thus the users can always choose which of the three differently generated binding to use. The binding site with the most similar experimentally observed knockdown percentage is given out, together with its properties and oligos ready to clone into the [https://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit miTuner]-construct.<br> | ||
Revision as of 01:24, 26 October 2010

|
|
||||||||||||
 "
"