Team:Groningen/12 July 2010
From 2010.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Groningen/Header}} | {{Team:Groningen/Header}} | ||
| - | '''Week 28, Arend Jan''' | + | '''Week 28''' |
| + | ---- | ||
| + | '''Laura''' | ||
| + | Reading articles about biofilm modeling, search for more info. | ||
| + | |||
| + | |||
| + | '''Arend Jan''' | ||
| Line 38: | Line 44: | ||
The transformation did not work. The pNZ8048 plasmid was isolated from E.coli NM522 and the ligation was trasformed to TOP10. Although the strains have the same methylation enzymes the positive control transformation also did not work. The only difference is that NM522 is EndA+ and TOP10 is EndA- but that should not matter. | The transformation did not work. The pNZ8048 plasmid was isolated from E.coli NM522 and the ligation was trasformed to TOP10. Although the strains have the same methylation enzymes the positive control transformation also did not work. The only difference is that NM522 is EndA+ and TOP10 is EndA- but that should not matter. | ||
We will need to design primers to get rid of the IS element. | We will need to design primers to get rid of the IS element. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | '''Geeske''' | ||
| + | |||
| + | <br> | ||
| + | Two failed attempts at transforming ''Bacillus subtilis'' NZ8900 (B. sub 168 with pNZ8900 integrated in the genome) with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3) (produced bij Arend Jan as stated above). Transformation performed according to protocol listed on protocol page. Exception from protocol: No FeNH4-citrate added to medium. | ||
| + | |||
| + | <br> | ||
'''Peter''' | '''Peter''' | ||
Latest revision as of 21:55, 25 October 2010
Week 28
Laura Reading articles about biofilm modeling, search for more info.
Arend Jan
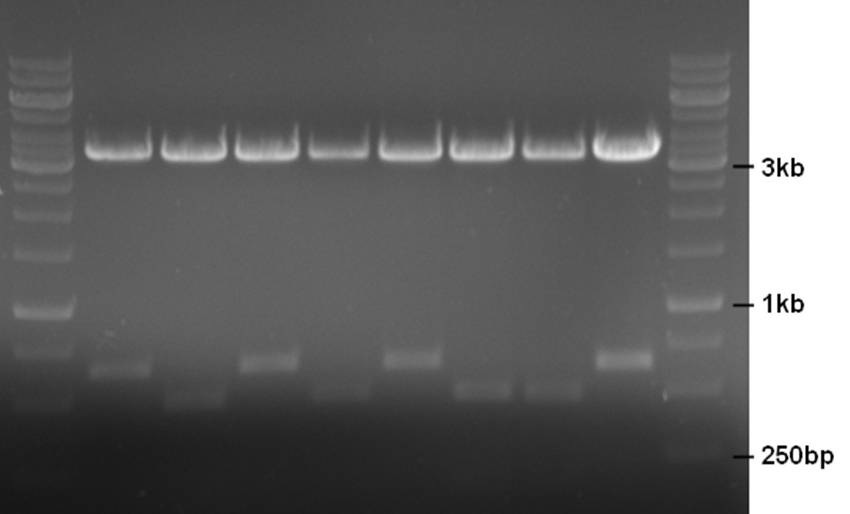
A mix of the ChpE and ChpH genes were ligated into the pNZ8901-bbs backbone and a restriction check was performed on 8 clones. ChpE contains a DpnI site whereas ChpH does not. A restriction with DpnI and XhoI should yield a 487bp and 176bp fragment for ChpE and just a 648bp fragment for ChpH (+ a ~3500bp backbone fragment for both).
- 10ul plasmid - 1.5ul buffer R - 0.5ul DpnI - 0.5ul XhoI - 2.5ul MQ
Clones 2, 4, 6, and 7 contain ChpE, and 1, 3, 5, and 8 contain ChpH. Clones 1, 2, 3, and 6 were sent for sequencing.
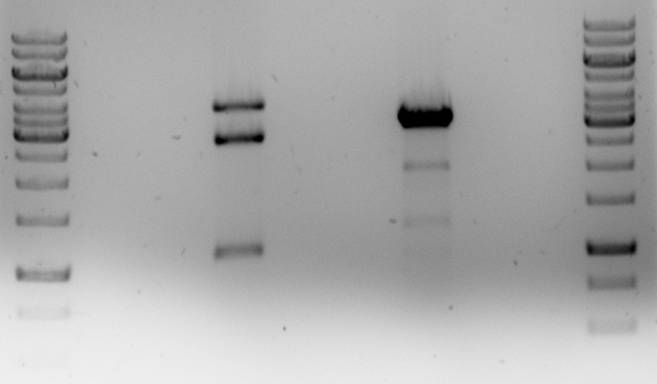
These ligations were performed with the backbone that still contains the unwanted IS element with the PstI site. At the same time we are attempting to delete this element using the related plasmid pNZ8048. This plasmid is the same as pNZ8901 except that it has a nisin inducible promoter. A restriction of the two plasmids with XhoI and MunI and combining two of their fragments would produce the pNZ8901-bbs plamid without the IS element.
In both cases the plasmid was only partially digested. The pNZ8901-bbs restriction did give an excisable fragment (~1100bp).
The pNZ8084 restriction was repeated.
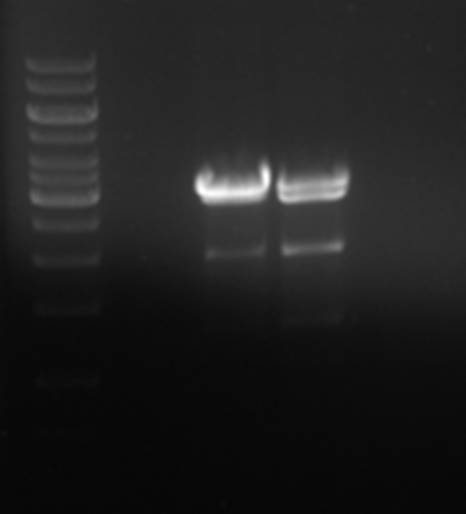
Again, for some reason, the plasmid was only partially digested, but the desired ~2000bp fragment is present. The fragment was isolated from gel and ligated with the pNZ8901-bbs fragment.
The transformation did not work. The pNZ8048 plasmid was isolated from E.coli NM522 and the ligation was trasformed to TOP10. Although the strains have the same methylation enzymes the positive control transformation also did not work. The only difference is that NM522 is EndA+ and TOP10 is EndA- but that should not matter.
We will need to design primers to get rid of the IS element.
Geeske
Two failed attempts at transforming Bacillus subtilis NZ8900 (B. sub 168 with pNZ8900 integrated in the genome) with pNZ8901_E (from clones 2 and 6) and pNZ8901_H (from clones 1 and 3) (produced bij Arend Jan as stated above). Transformation performed according to protocol listed on protocol page. Exception from protocol: No FeNH4-citrate added to medium.
Peter
A degredation experiment was performed, to test whether the chaplins were degraded.
This degredation experiment was performed according to protocol (DegredationGR), using all strains of B. subtilis and purified chaplins from Streptomyces.
Analysis was done by SDS-Page.
Results
 "
"