Team:WashU/Yeast
From 2010.igem.org
BrianLandry (Talk | contribs) (→Chromosomal Integration) |
BrianLandry (Talk | contribs) (→Yeast Vectors) |
||
| Line 2: | Line 2: | ||
==Background== | ==Background== | ||
===Yeast Vectors=== | ===Yeast Vectors=== | ||
| - | Currently there exist two different methods for insertion of synthetic DNA into yeast cells, plasmid vectors or chromosomal integration. Plasmid vectors contain the components necessary to replicate autonomously after entering the yeast cells. Chromosomal integration inserts DNA directly into the yeast chromosome | + | Currently there exist two different methods for insertion of synthetic DNA into yeast cells, plasmid vectors or chromosomal integration. Plasmid vectors contain the components necessary to replicate autonomously after entering the yeast cells. Chromosomal integration inserts DNA directly into the yeast chromosome from a linear construct. |
====Yeast Plasmids ==== | ====Yeast Plasmids ==== | ||
| - | There exist two main types of plasmids used to insert and maintain DNA in yeast cells, Yeast Episomal plasmids (YEp) and Yeast Centromer containing plasmids (YCp). Natively yeast contain a plasmid referred to as the 2 μm plasmid, YEp vectors use an origin of replication derived from this plasmid and maintain copy numbers of 50-100. The YCp contain an Autonomous Replicating Sequence (ARS) and a yeast Centromer sequence (CEN) and maintain a copy number of 1-3 per cell | + | There exist two main types of plasmids used to insert and maintain DNA in yeast cells, Yeast Episomal plasmids (YEp) and Yeast Centromer containing plasmids (YCp). Natively yeast contain a plasmid referred to as the 2 μm plasmid, YEp vectors use an origin of replication derived from this plasmid and maintain copy numbers of 50-100. The YCp contain an Autonomous Replicating Sequence (ARS) and a yeast Centromer sequence (CEN) and maintain a copy number of 1-3 per cell (Keshav). |
Commonly these plasmids will also contain an E. coli origin of replication and selectable marker, this allows for DNA manipulation in E. coli, and then the transfer of the plasmid directly into yeast, earning this type of plasmid the name "Shuttle Vectors". | Commonly these plasmids will also contain an E. coli origin of replication and selectable marker, this allows for DNA manipulation in E. coli, and then the transfer of the plasmid directly into yeast, earning this type of plasmid the name "Shuttle Vectors". | ||
Revision as of 21:00, 25 October 2010

Background
Yeast Vectors
Currently there exist two different methods for insertion of synthetic DNA into yeast cells, plasmid vectors or chromosomal integration. Plasmid vectors contain the components necessary to replicate autonomously after entering the yeast cells. Chromosomal integration inserts DNA directly into the yeast chromosome from a linear construct.
Yeast Plasmids
There exist two main types of plasmids used to insert and maintain DNA in yeast cells, Yeast Episomal plasmids (YEp) and Yeast Centromer containing plasmids (YCp). Natively yeast contain a plasmid referred to as the 2 μm plasmid, YEp vectors use an origin of replication derived from this plasmid and maintain copy numbers of 50-100. The YCp contain an Autonomous Replicating Sequence (ARS) and a yeast Centromer sequence (CEN) and maintain a copy number of 1-3 per cell (Keshav).
Commonly these plasmids will also contain an E. coli origin of replication and selectable marker, this allows for DNA manipulation in E. coli, and then the transfer of the plasmid directly into yeast, earning this type of plasmid the name "Shuttle Vectors".
However, the use of these plasmids is not without drawbacks. Both types of origins face significance loss of plasmid on the order of 10-2. When using the 2 μm replication origin and grown in selective media 60% to 95% of yeast cells may not contain the plasmid (Sherman). Additionally the variability in copy number can introduces noise when quantify the amount of protein expressed in cells.
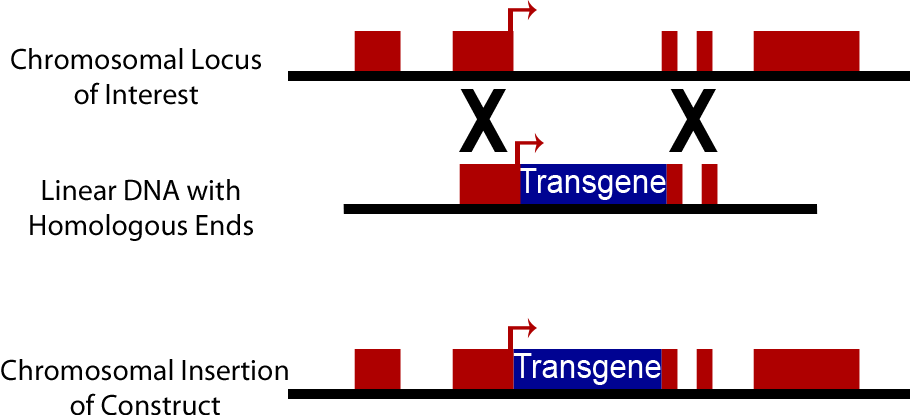
Chromosomal Integration
An alternative to the use of plasmids in yeast is direct integration into the chromosome of yeast cells. Yeast cells natively have high levels of homologous recombination activity, consequently when a DNA sequence is introduced into a yeast cell containing sequences homologous to those present on the yeast chromosome, it can be swapped into the yeast chromosome. This method is commonly used to create knock out strains excising genes such as Ura3, His3, Trp1, His3 and Lys2. It can also be used to insert synthetic DNA constructs into the genome. Chromosomally integrated DNA is lost at a much lower rate, approximately 10^-3 to 10^-4, than plasmid discharge (Sherman). However transformation efficiency is lower than with plasmids, on an order of 10^3 transformants/μg opposed to efficiency of 10^5 transformants/μg when plasmids are used. Yeast Integrating plasmids (YIp) contain homologous sequences to the yeast chromosome and cause integration into the yeast chromosome. To increase transformation efficiency by 10^2 these plasmids are linearized before transformation (Keshav).
Yeast Selection
Project Goals
Yeast Vectors
Two new plasmid backbones are to be created and submitted to the registry. Similar in structure the two vectors will be derived from biobrick [http://partsregistry.org/wiki/index.php?title=Part:pSB1AT3 pSB1AT3] which contains a high copy E. coli replication origin and both ampicillin and tetracycline resistance. PCR will be use to modify the site upstream of the prefix and downstream of the suffix in vector. The Plasmid will take the following design:
This allows the insertion of yeast biobrick parts (including positive selection markers) into the vector using E. coli. Once the construct is fully assembled the vector is linearized with BbsI and transformed into yeast. BbsI was chosen since it cuts downstream of its recognition site, and will create non-overlapping ends preventing circularization of the linear products. The two new vectors will contain sequences homologous to the Ura3 and His3 loci in the yeast genome. When employed either the Ura3 or His3 loci will be replaced with the construct, creating an autotrophic strain of yeast. When coupled with a positive selection marker this will allow for both positive and negative selection of transfomants.
Positive Selection Markers
Results
Works Cited
S. Keshav, J. Heinemann. Yeast Plasmids, 1997 v.62, 113-130
An Introduction to the Genetics and Molecular Biology of the Yeast Saccharomyces cerevisiae. F. Sherman, 1998. http://dbb.urmc.rochester.edu/labs/sherman_f/yeast/Index.html
Yeast Protocols Handbook. Clontech, 1990
 "
"