Team:LMU-Munich/Cut'N'survive/Schedule
From 2010.igem.org
| (30 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| + | |||
| + | [[Image:cnsZelle.png|300px|Logo|right]] | ||
| + | |||
| + | |||
| + | |||
==Aim 1: DNA replication+PCR== | ==Aim 1: DNA replication+PCR== | ||
For PCR key see [[Team:LMU-Munich/Cut'N'survive/Schedule/PCR key | PCR key]] | For PCR key see [[Team:LMU-Munich/Cut'N'survive/Schedule/PCR key | PCR key]] | ||
| + | |||
<b>Colourcode for the primers: <font color="#FF0000">Annealing part</font>, <font color="#009933">mutagenised part</font>, <font color="#FF6600">other sequences integrated into primer</font></b> | <b>Colourcode for the primers: <font color="#FF0000">Annealing part</font>, <font color="#009933">mutagenised part</font>, <font color="#FF6600">other sequences integrated into primer</font></b> | ||
| - | PCR1: replication of the Tet-inducible CMV promotor [X] | + | |
| + | <b>PCR1: replication of the Tet-inducible CMV promotor [X]</b> | ||
[[Image:PCR1.jpg]] | [[Image:PCR1.jpg]] | ||
| - | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer1| 1TreF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer2| 2TreR]] | + | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer1| 1TreF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer2| 2TreR]] |
| + | expected product size: 492bp | ||
| - | PCR2a: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [ ] | + | |
| + | <b>PCR2a: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [X]</b> | ||
[[Image:PCR2a.jpg]] | [[Image:PCR2a.jpg]] | ||
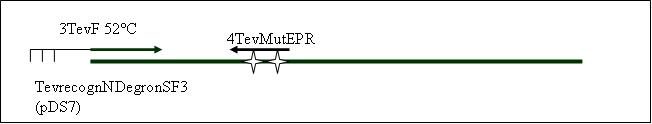
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer3| 3TevF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer4| 4TevMutEPR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer3| 3TevF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer4| 4TevMutEPR]] | ||
| + | expected product size: 332 bp | ||
| + | |||
| - | PCR2b: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [ ] | + | <b>PCR2b: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [X]</b> |
[[Image:PCR2b.jpg]] | [[Image:PCR2b.jpg]] | ||
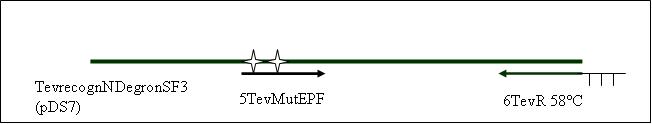
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer5| 5TevMutEPF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer6| 6TevR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer5| 5TevMutEPF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer6| 6TevR]] | ||
| + | expected product size: 772 bp | ||
| - | PCR3: joining PCR of the TEVrecogn-NDegron-SF3 Part [ ] | + | |
| + | <b>PCR3: joining PCR of the TEVrecogn-NDegron-SF3 Part [X]</b> | ||
[[Image:PCR3.jpg]] | [[Image:PCR3.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer3| 3TevF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer6| 6TevR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer3| 3TevF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer6| 6TevR]] | ||
| + | expected product size: 1087 bp | ||
| - | PCR4a: replication human bak with mutagenisis (Pst1) [ ] | + | |
| + | <b>PCR4a: replication human bak with mutagenisis (Pst1) [X]</b> | ||
[[Image:PCR4a.jpg]] | [[Image:PCR4a.jpg]] | ||
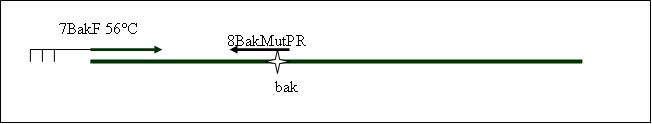
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer7| 7BakF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer8| 8BakMutPR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer7| 7BakF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer8| 8BakMutPR]] | ||
| + | expected product size: 330 bp | ||
| - | PCR4b: replication human bak with mutagenisis (Pst1) [ ] | + | |
| + | <b>PCR4b: replication human bak with mutagenisis (Pst1) [X]</b> | ||
[[Image:PCR4b.jpg]] | [[Image:PCR4b.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer9| 9BakMutPF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer10| 10BakR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer9| 9BakMutPF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer10| 10BakR]] | ||
| + | expected product size: 376 bp | ||
| + | |||
| - | PCR5: joining PCR of human bak [ ] | + | <b>PCR5: joining PCR of human bak [X]</b> |
[[Image:PCR5.jpg]] | [[Image:PCR5.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer7| 7BakF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer10| 10BakR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer7| 7BakF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer10| 10BakR]] | ||
| + | expected product size: 688 bp | ||
| - | PCR6: replication of the SV40-polyadenylation site [X] | + | |
| + | <b>PCR6: replication of the SV40-polyadenylation site [X]</b> | ||
[[Image:PCR6.jpg]] | [[Image:PCR6.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer11| 11PAF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer12| 12PAR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer11| 11PAF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer12| 12PAR]] | ||
| + | expected product size: 237 bp | ||
| - | PCR7a: replication of the p14*TEVrecogn with mutagenisis (Spe1) [ ] | + | |
| + | <b>PCR7a: replication of the p14*TEVrecogn with mutagenisis (Spe1) [X]</b> | ||
[[Image:PCR7a.jpg]] | [[Image:PCR7a.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer13| 13TrecF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer14| 14TrecMutSR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer13| 13TrecF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer14| 14TrecMutSR]] | ||
| + | expected product size: 850 bp | ||
| + | |||
| - | PCR7b: replication of the p14*TEVrecogn with mutagenisis (Spe1) with TEV recogn in primer (16TrecR) [ ] | + | <b>PCR7b: replication of the p14*TEVrecogn with mutagenisis (Spe1) with TEV recogn in primer (16TrecR) [X]</b> |
[[Image:PCR7b.jpg]] | [[Image:PCR7b.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer15| 15TrecMutSF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer16| 16TrecR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer15| 15TrecMutSF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer16| 16TrecR]] | ||
| + | expected product size: 402 bp | ||
| - | PCR8: joining PCR of p14*TEVrecogn with TEV recogn in primer (16TrecR) [ ] | + | |
| + | <b>PCR8: joining PCR of p14*TEVrecogn with TEV recogn in primer (16TrecR) [X]</b> | ||
[[Image:PCR8.jpg]] | [[Image:PCR8.jpg]] | ||
Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer13| 13TrecF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer16| 16TrecR]] | Primer used:[[Team:LMU-Munich/Cut'N'survive/Schedule/Primer13| 13TrecF]][[Team:LMU-Munich/Cut'N'survive/Schedule/Primer16| 16TrecR]] | ||
| + | expected product size: 1233 bp | ||
| + | |||
| + | ==Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequence == | ||
| + | |||
| + | PCR1 (BBa_K368001): Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR1| Sequence of PCR1 from sequencing]]: confirmed [ ] | ||
| + | |||
| + | PCR3 (BBa_K368016): Insertion [X] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR3| Sequence of PCR3 from sequencing]]: confirmed [X] | ||
| + | |||
| + | PCR5 (BBa_K368017): Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR5| Sequence of PCR5 from sequencing]]: confirmed [ ] | ||
| + | |||
| + | PCR6 (BBa_K368018): Insertion [ ] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR6| Sequence of PCR6 from sequencing]]: confirmed [ ] | ||
| + | |||
| + | PCR8 (BBa_K368019): Insertion [X] -> [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzPCR8| Sequence of PCR8 from sequencing]]: confirmed [X] | ||
| + | |||
| + | ==Aim 3: Assembling Biobricks== | ||
| + | |||
| + | We are using the [[Team:LMU-Munich/Notebook/Protocols/13_3A_Method_for_Biobrick_assembly|3A System]] to assemble Biobricks. | ||
| + | |||
| + | Assembled Biobricks: | ||
| + | |||
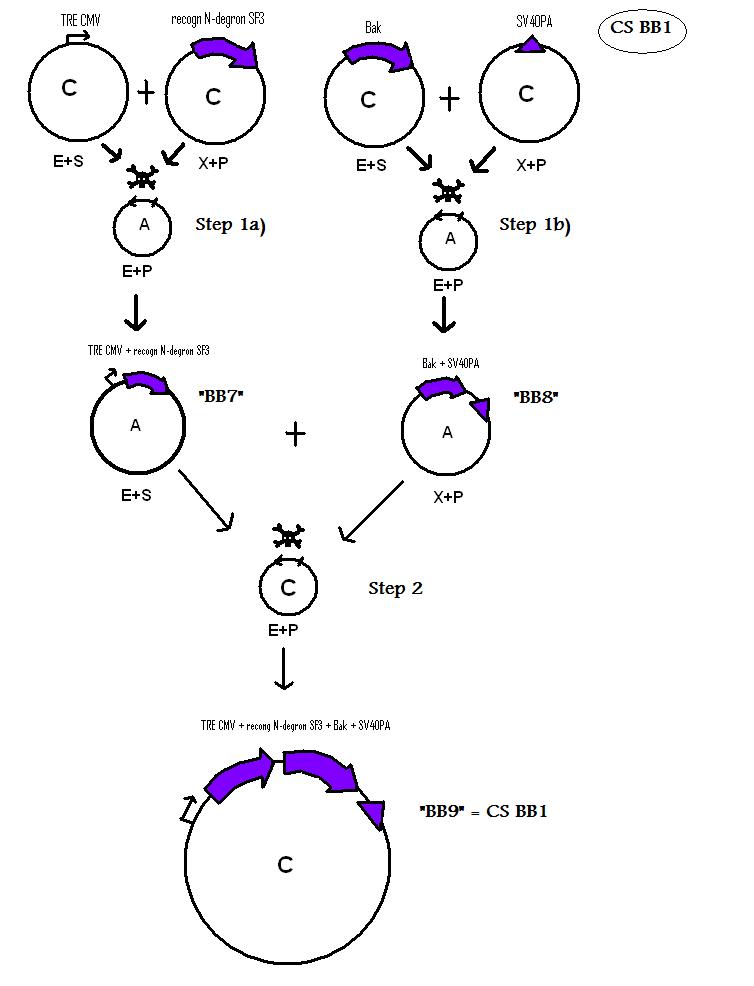
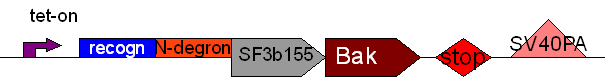
| + | * BB7 (BBa_K368007): assembled [ ]; tet-on promoter (prev TRE CMV) + TEV recognition site + SF3b155; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB7| Sequence of BB7 from sequencing]] confirmed: [ ] | ||
| + | |||
| + | * BB8 (BBa_K368008): assembled [ ]; Human Bak + SV40PA ; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB8| Sequence of BB8 from sequencing]] confirmed: [ ] | ||
| + | |||
| + | * BB9 [BBa_K368009): assembled [ ]; tet-on promoter (prev TRE CMV) + TEV recognition site + SF3b155 + Human Bak + SV40PA; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB9| Sequence of BB9 from sequencing]] confirmed: [ ] | ||
| + | |||
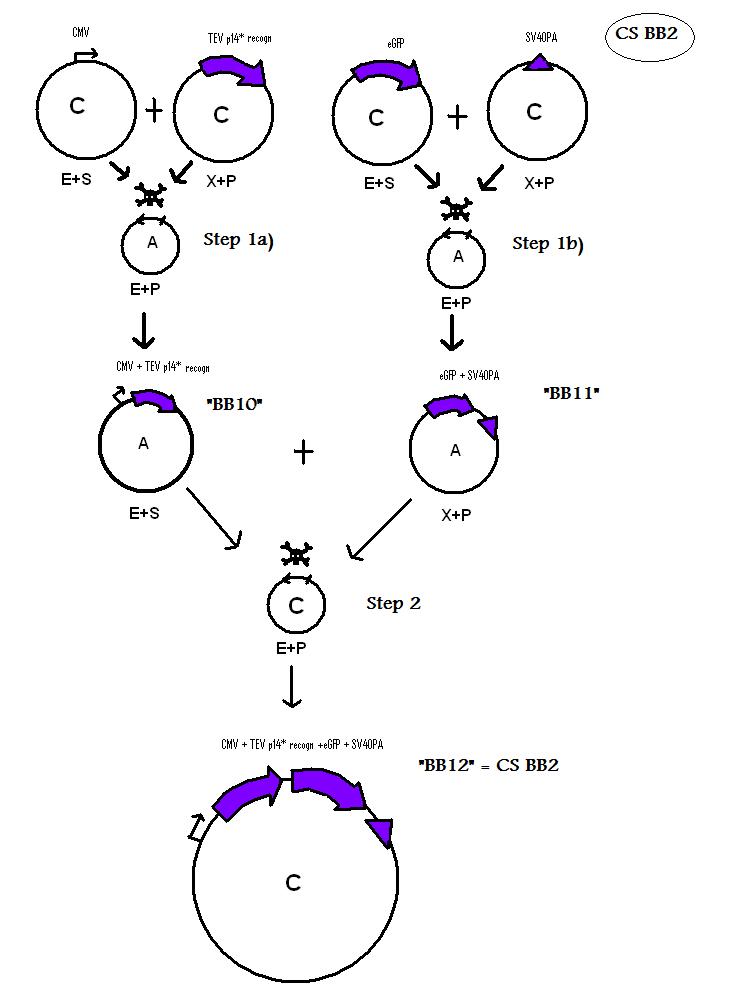
| + | * BB10 (BBa_K368010): assembled [ ]; CMV-promoter + TEV protesase + p14* + TEV recognition site ; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB10| Sequence of BB10 from sequencing]] confirmed: [ ] | ||
| + | |||
| + | * BB11 (BBa_K368011): assembled [X]; eGFP + SV40PA ; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB11| Sequence of BB11 from sequencing]] confirmed: [X] | ||
| + | |||
| + | * BB12 (BBa_K368012): assembled [ ]; CMV-promoter + TEV protesase + p14* + TEV recognition site + eGFP + SV40PA; | ||
| + | |||
| + | [[Team:LMU-Munich/Jump-or-die/Schedule/SequenzBB12| Sequence of BB12 from sequencing]] confirmed: [ ] | ||
| + | |||
| - | + | <b> Assembling Construct 1 </b> | |
| - | + | ||
| - | + | ||
| + | [[Image: N1S.jpg]] | ||
| + | <b> Assembling Construct 2 </b> | ||
| - | + | [[Image: N2S.jpg]] | |
| - | + | ==Aim 4: Testing products== | |
| - | + | === Construct 1 === | |
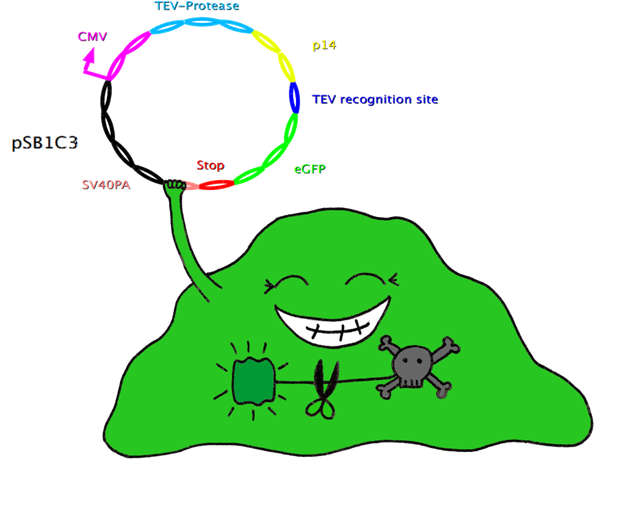
| - | [[ | + | [[Image:Konstruktcut1.png|400pxs|TEV Construct 1: Inserted in celline]] |
| - | + | - transform into HeLa cells to see if they survive. | |
| + | -> Check the leakiness of tet-on-promoter | ||
| + | - induce tet-on-promoter to see, if cells die. | ||
| - | + | -> Check if construct 1 is working | |
| - | + | === Construct 2 === | |
| - | + | [[Image:Konstruktcut2.png|400pxs|TEV Construct 2: Contains gene of interest]] | |
| - | + | ||
| - | + | - Transform into HeLa cells and see if eGFP is being translated | |
| - | + | -> Check if CMV-promoter is working and construct is being read-off completely | |
| - | + | - Western Blot | |
| - | + | -> Check by reference of protein size if eGFP is cut off from the residual of the protein by TEV-Protease | |
| - | + | ==Aim 5: Testing system== | |
| - | |||
| - | |||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 14:32, 25 October 2010
For PCR key see PCR key
Colourcode for the primers: Annealing part, mutagenised part, other sequences integrated into primer
Primer used: 1TreF 2TreR
expected product size: 492bp
Primer used: 3TevF 4TevMutEPR
expected product size: 332 bp
Primer used: 5TevMutEPF 6TevR
expected product size: 772 bp
Primer used: 3TevF 6TevR
expected product size: 1087 bp
Primer used: 7BakF 8BakMutPR
expected product size: 330 bp
Primer used: 9BakMutPF 10BakR
expected product size: 376 bp
Primer used: 7BakF 10BakR
expected product size: 688 bp
Primer used: 11PAF 12PAR
expected product size: 237 bp
Primer used: 13TrecF 14TrecMutSR
expected product size: 850 bp
Primer used: 15TrecMutSF 16TrecR
expected product size: 402 bp
Primer used: 13TrecF 16TrecR
expected product size: 1233 bp
PCR1 (BBa_K368001): Insertion [ ] -> Sequence of PCR1 from sequencing: confirmed [ ]
PCR3 (BBa_K368016): Insertion [X] -> Sequence of PCR3 from sequencing: confirmed [X]
PCR5 (BBa_K368017): Insertion [ ] -> Sequence of PCR5 from sequencing: confirmed [ ]
PCR6 (BBa_K368018): Insertion [ ] -> Sequence of PCR6 from sequencing: confirmed [ ]
PCR8 (BBa_K368019): Insertion [X] -> Sequence of PCR8 from sequencing: confirmed [X]
We are using the 3A System to assemble Biobricks.
Assembled Biobricks:
Sequence of BB7 from sequencing confirmed: [ ]
Sequence of BB8 from sequencing confirmed: [ ]
Sequence of BB9 from sequencing confirmed: [ ]
Sequence of BB10 from sequencing confirmed: [ ]
Sequence of BB11 from sequencing confirmed: [X]
Sequence of BB12 from sequencing confirmed: [ ]
Assembling Construct 2
- transform into HeLa cells to see if they survive.
-> Check the leakiness of tet-on-promoter
- induce tet-on-promoter to see, if cells die.
-> Check if construct 1 is working
- Transform into HeLa cells and see if eGFP is being translated
-> Check if CMV-promoter is working and construct is being read-off completely
- Western Blot
-> Check by reference of protein size if eGFP is cut off from the residual of the protein by TEV-Protease


![]()
![]()







![]()
![]()

![]()
Contents
Aim 1: DNA replication+PCR
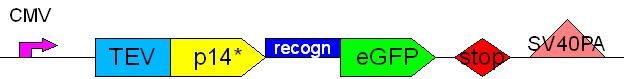
PCR1: replication of the Tet-inducible CMV promotor [X]

PCR2a: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [X]
PCR2b: replication of the TEVrecogn-NDegron-SF3 Part with mutagenisis (EcoR1 + Pst1) [X]
PCR3: joining PCR of the TEVrecogn-NDegron-SF3 Part [X]

PCR4a: replication human bak with mutagenisis (Pst1) [X]
PCR4b: replication human bak with mutagenisis (Pst1) [X]
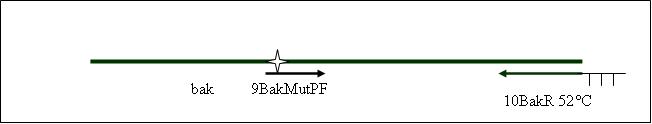
PCR5: joining PCR of human bak [X]
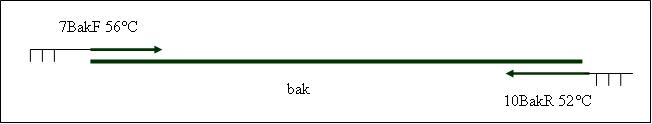
PCR6: replication of the SV40-polyadenylation site [X]

PCR7a: replication of the p14*TEVrecogn with mutagenisis (Spe1) [X]
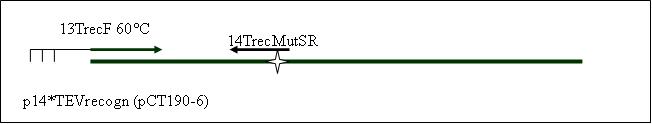
PCR7b: replication of the p14*TEVrecogn with mutagenisis (Spe1) with TEV recogn in primer (16TrecR) [X]
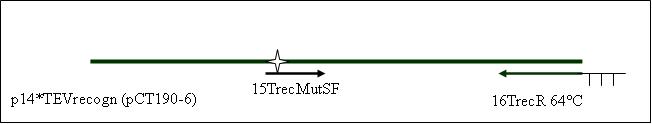
PCR8: joining PCR of p14*TEVrecogn with TEV recogn in primer (16TrecR) [X]

Aim 2: Inserting PCR Products in pSB1C3 and verifying Sequence
Aim 3: Assembling Biobricks
Assembling Construct 1
Aim 4: Testing products
Construct 1
Construct 2
Aim 5: Testing system
![]()
![]()

 "
"