Team:TU Delft/Project/alkane-degradation/results
From 2010.igem.org
(Difference between revisions)
(→Alkane Degradation Results & Conclusions) |
(→Alkb2 Cluster) |
||
| Line 5: | Line 5: | ||
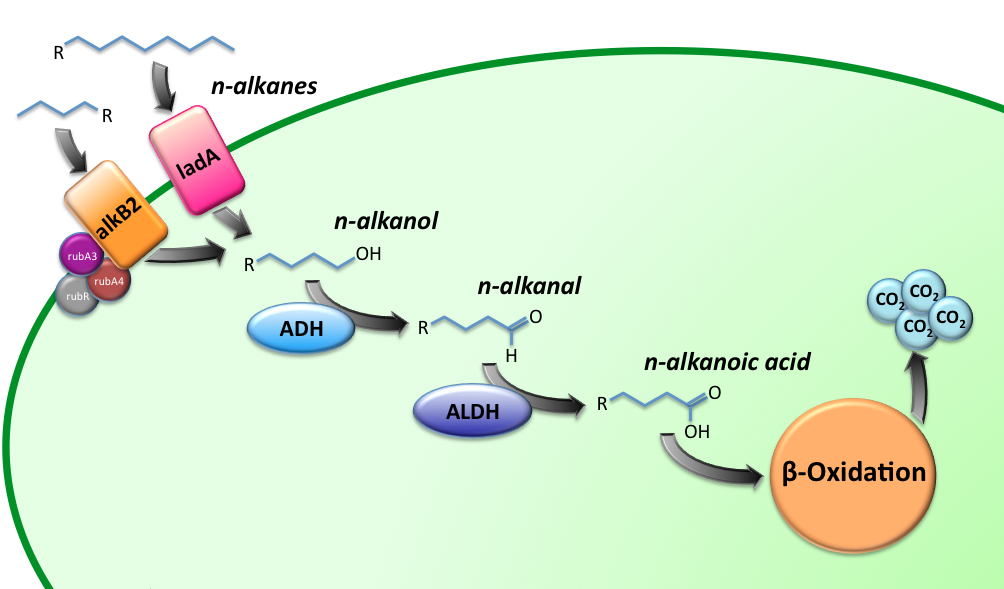
[[Image:TUDelft_Alkane_degradation_route.png|600px|thumb|center|'''Figure 1''' – Schematic description of the alkane degradation pathway with the corresponding genes.]] | [[Image:TUDelft_Alkane_degradation_route.png|600px|thumb|center|'''Figure 1''' – Schematic description of the alkane degradation pathway with the corresponding genes.]] | ||
| - | === | + | ===Alkane hydroxylase (AH) system=== |
===LadA=== | ===LadA=== | ||
Revision as of 20:11, 24 October 2010

Alkane Degradation Results & Conclusions
Alkane hydroxylase (AH) system
LadA
ADH
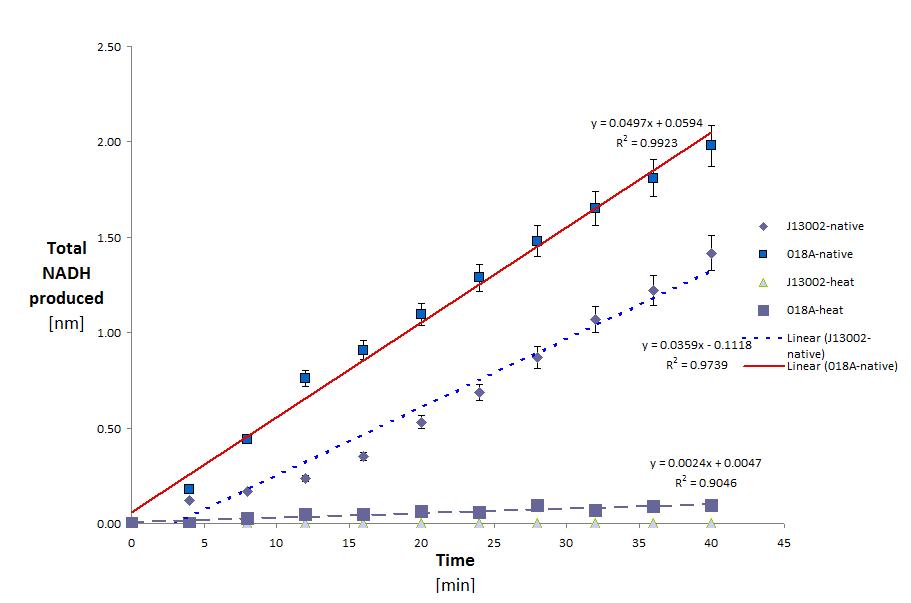
ADH was tested with Dodecanol (C12)
This yielded the following results:
| Conditions | Native | Native | Heated | Heated |
| Sample | J13002 | K398018 | J13002 | K398018 |
| Total protein in well | 3.303E-02 | 3.180E-02 | 7.351E-03 | 7.994E-03 |
| Specific activity (kat/mg) | 1.812E-11 | 2.606E-11 | 0.000E-00 | 5.041E-12 |
This shows two things. That E.Coli is partly able to degrade Dodecanol on it's own. And that this activity is greatly increased (43.83% normalized for the amount of protein) with the addition of the ADH gene. It is also clear that eventhough ADH is from a thermophilic organism it loses most (63.5%) of it's activity after heating. the raw data for these calculations can be seen below. (Although this decrease in activity could also be attributed to the host organism lacking the proper post translational modifications.)
ALDH

 "
"