Team:UNIPV-Pavia/Calendar/August/settimana2
From 2010.igem.org
m (→August, 10th) |
m (→August, 10th) |
||
| (88 intermediate revisions not shown) | |||
| Line 30: | Line 30: | ||
<html><p align="center"><font size="4"><b>AUGUST: WEEK 2</b></font></p></html><hr><br> | <html><p align="center"><font size="4"><b>AUGUST: WEEK 2</b></font></p></html><hr><br> | ||
| + | <html><a name="indice"/></html> | ||
==August, 9th== | ==August, 9th== | ||
Phasins plates showed very few colonies (12 for I20-new and 17 for I21-new): they were all picked and let grow in LB+Amp 100ug/ml. In the evening we made glycerol stocks and re-filled the tubes for the screening of the following day: | Phasins plates showed very few colonies (12 for I20-new and 17 for I21-new): they were all picked and let grow in LB+Amp 100ug/ml. In the evening we made glycerol stocks and re-filled the tubes for the screening of the following day: | ||
{|border="1" align="center" | {|border="1" align="center" | ||
| - | |'''I20-new:'''|| | + | |'''I20-new:'''||I20-1||I20-2||I20-3||I20-4||I20-5||I20-6||I20-7||I20-8||I20-9||I20-10||I20-11||I20-12 |
|- | |- | ||
| - | |'''I21-new:'''|| | + | |'''I21-new:'''||I21-1||I21-2||I21-3||I21-4||I21-5||I21-6||I21-7||I21-8||I21-9||I21-10||I21-11||I21-12||I21-13||I21-14||I21-15||I21-16||I21-17 |
|} | |} | ||
In order not to loose our time we decided to perform again the PCR-amplification/modification of <partinfo>BBa_K208001</partinfo> in case of a negative screening. Than we gel-ran and gel-extracted right amplicons. | In order not to loose our time we decided to perform again the PCR-amplification/modification of <partinfo>BBa_K208001</partinfo> in case of a negative screening. Than we gel-ran and gel-extracted right amplicons. | ||
| Line 59: | Line 60: | ||
|I15-1 || 5ml LB+Amp || Ligation | |I15-1 || 5ml LB+Amp || Ligation | ||
|- | |- | ||
| - | | Linker || 5ml LB+Amp || Ligation | + | | Linker (<partinfo>BBa_K105012</partinfo>) || 5ml LB+Amp || Ligation |
|- | |- | ||
| - | | pSB4C5 || 5ml LB+Cm 12,5 || Ligation | + | | <partinfo>pSB4C5</partinfo> || 5ml LB+Cm 12,5 || Ligation |
|- | |- | ||
| I22-1 || 5ml LB+ Amp (picked from colony)|| Ligation/Screening/Glycerol stock | | I22-1 || 5ml LB+ Amp (picked from colony)|| Ligation/Screening/Glycerol stock | ||
| Line 102: | Line 103: | ||
*MC123/008 in LB+Amp 50 ug/ml | *MC123/008 in LB+Amp 50 ug/ml | ||
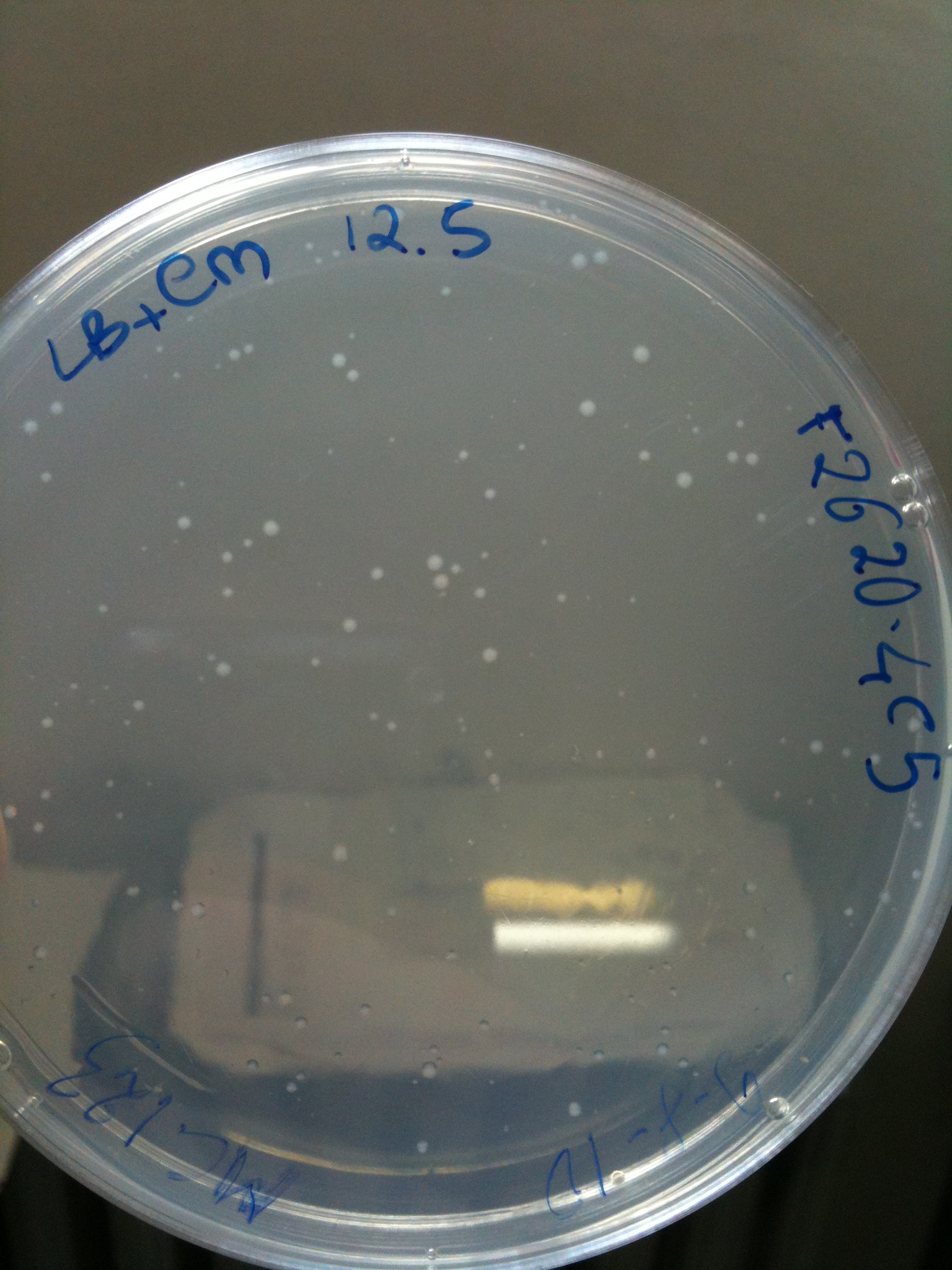
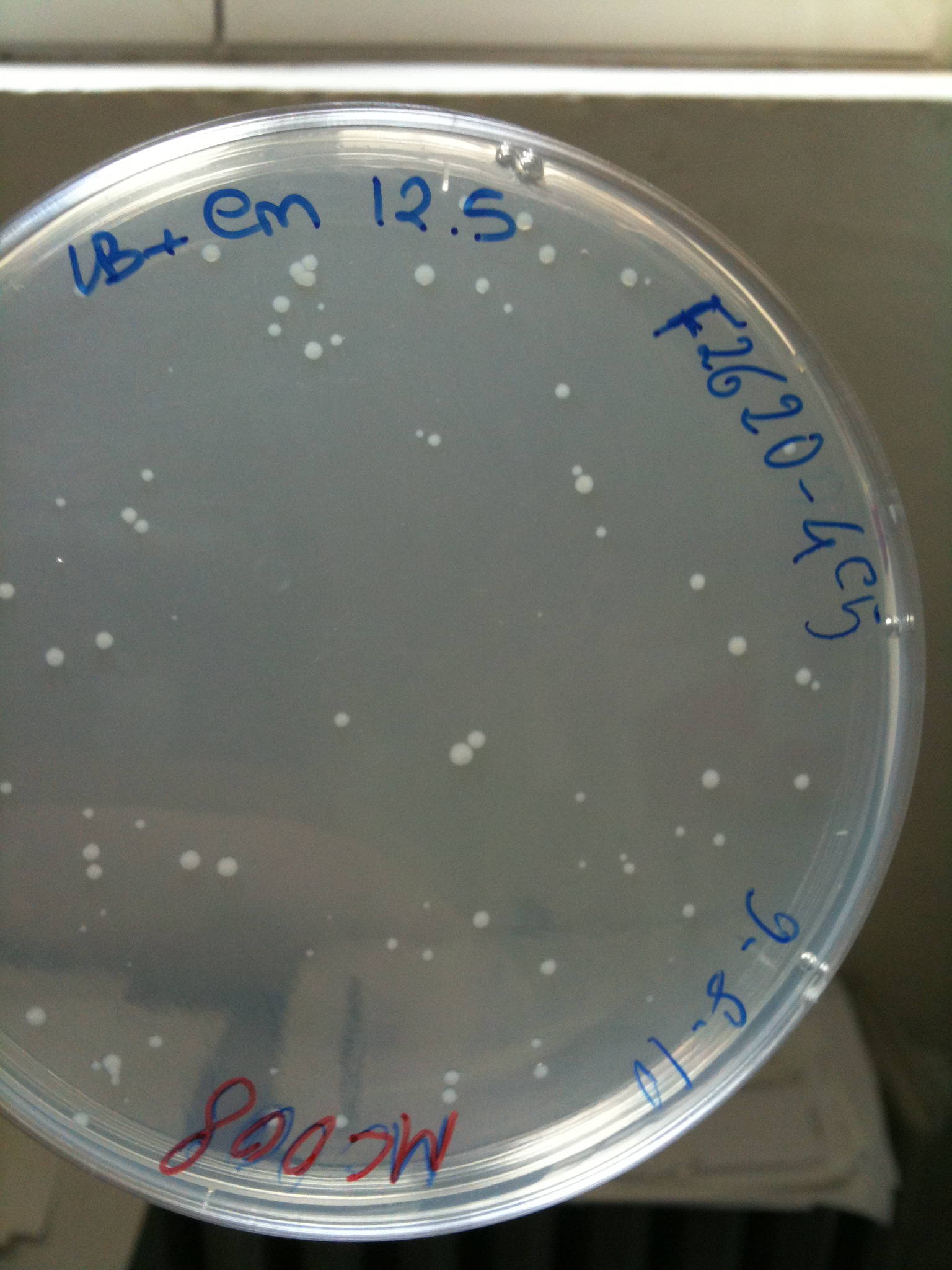
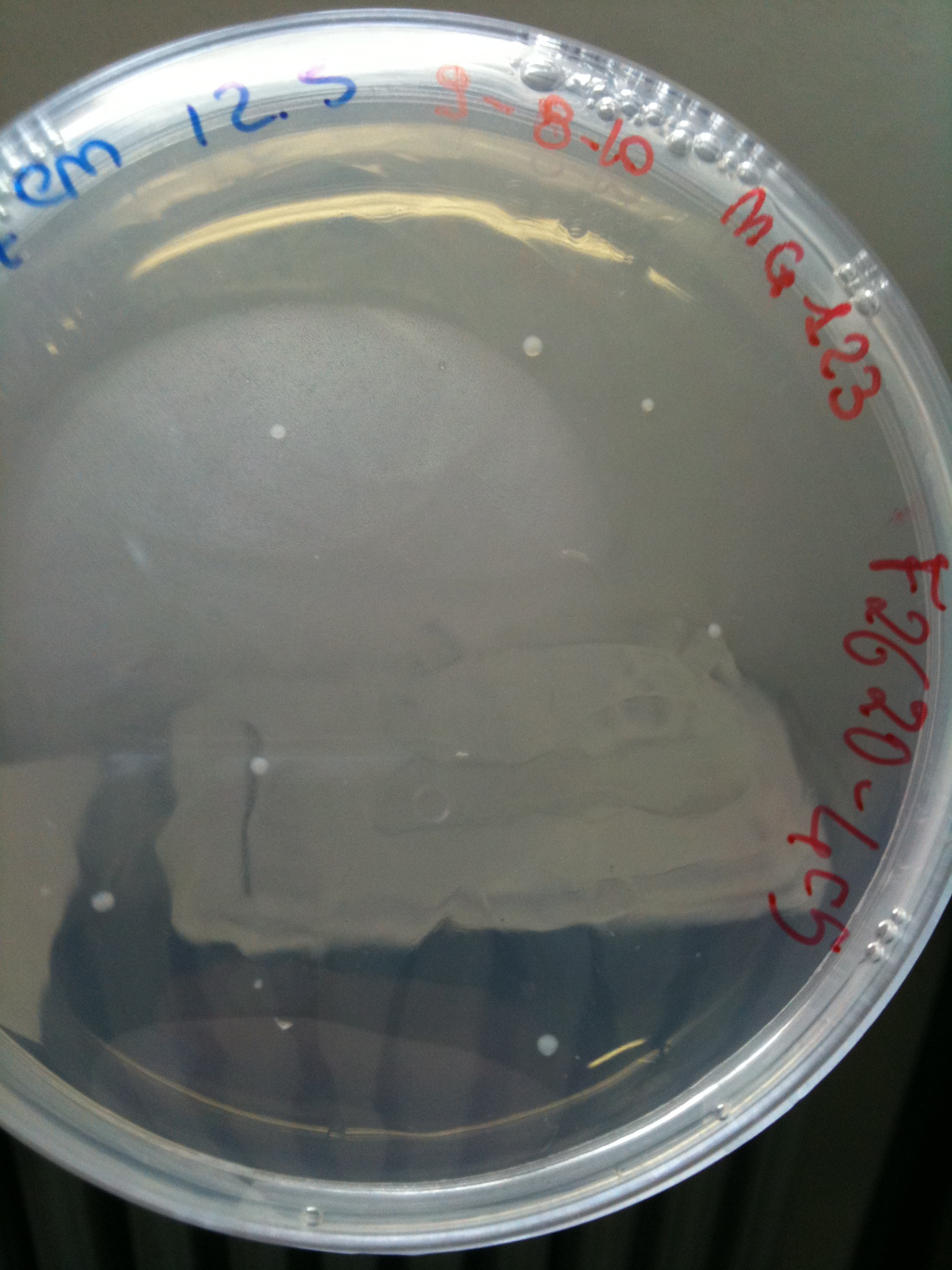
*F2620-4C5 (positive control) | *F2620-4C5 (positive control) | ||
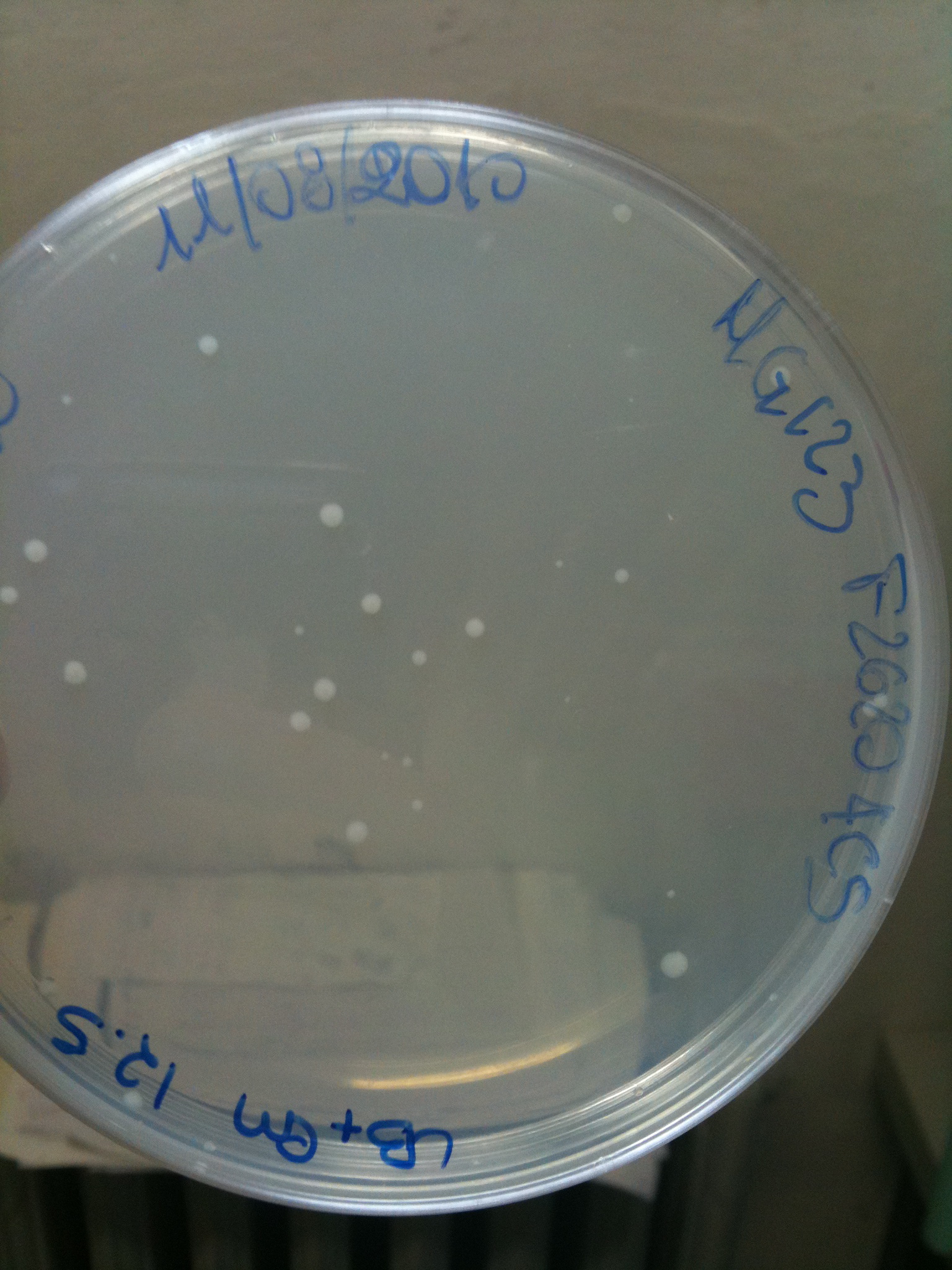
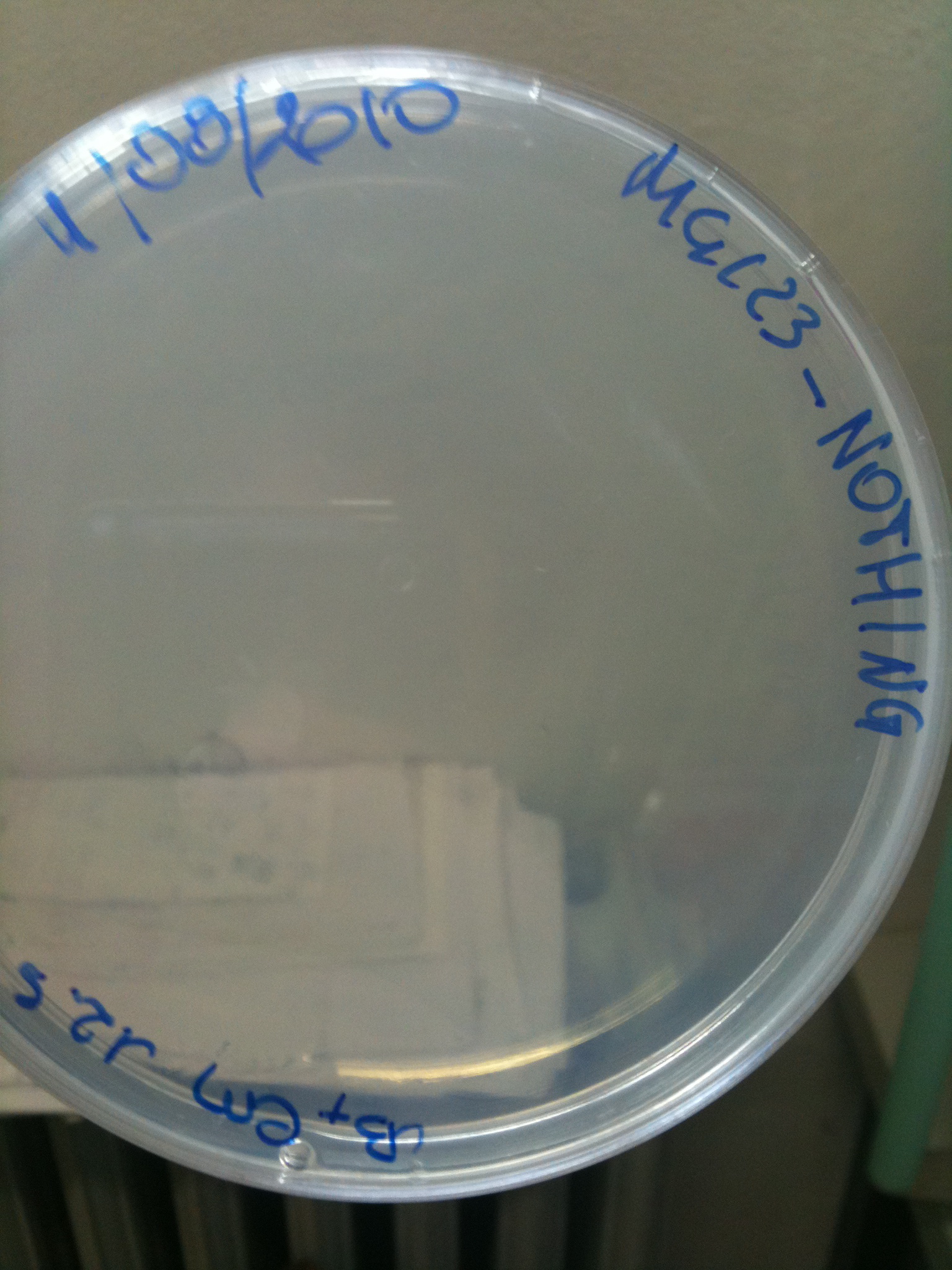
| - | Than we streaked cultures on a seven-sector divided plate, and we let | + | Than we streaked cultures on a seven-sector divided plate, and we let it grow ON at 30°C. Since 6 ug/ml is a very low concentration, we wanted to check if actually nothing (except F2620-4C5) grows on these plates. |
---- | ---- | ||
| - | Transformation at 30°C of 100 ul of | + | Transformation at 30°C of 100 ul of MG123/008 and MC123/008 competent cells to check their efficiency. We used 1 ul (~4 ng) of RING, F2620-4C5, NOTHING (water) and plated on proper LB agar plates: |
*RING: Cm 34 ug/ml | *RING: Cm 34 ug/ml | ||
*F2620-4C5 (positive control): Cm 12,5 ug/ml | *F2620-4C5 (positive control): Cm 12,5 ug/ml | ||
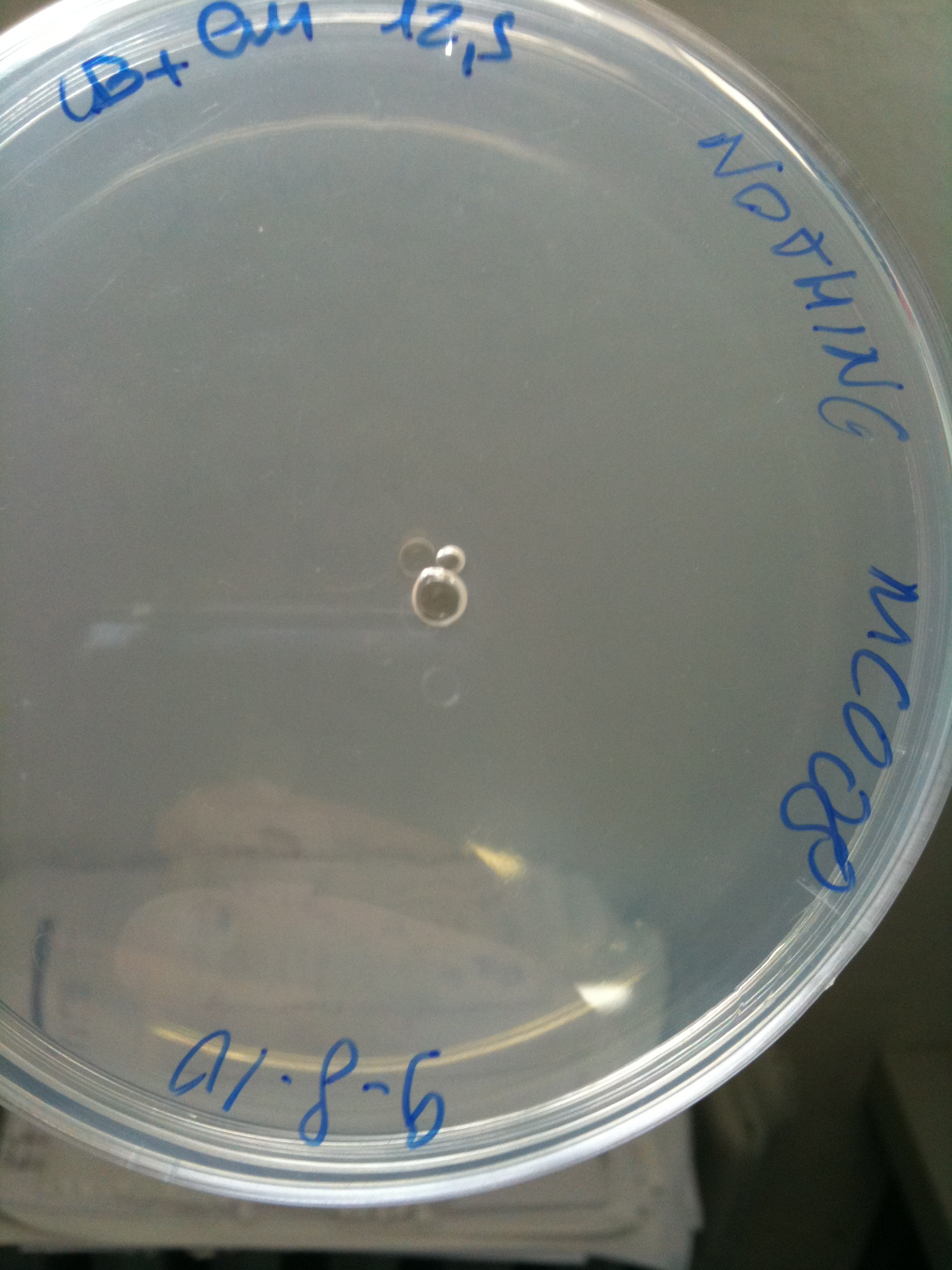
| - | * | + | *NOTHING (negative control): Cm 12,5 ug/ml |
| - | Both | + | Both F2620-4C5 and RING should survive (not RING when in XX123), but we had a problem so that MC008 transformed with RING couldn't be plated. We will check it another time. We let grow plates ON, 30°C. |
| - | + | <div align="right"><small>[[#indice|^top]]</small></div> | |
| - | + | ||
==August, 10th== | ==August, 10th== | ||
| Line 117: | Line 117: | ||
Glycerol stock was prepared for: | Glycerol stock was prepared for: | ||
| - | * linker | + | * linker (<partinfo>BBa_K105012</partinfo>) |
* I22-1 | * I22-1 | ||
* I22-2 | * I22-2 | ||
| Line 129: | Line 129: | ||
and are stored at -80°C. | and are stored at -80°C. | ||
| - | |||
---- | ---- | ||
| - | Cultures were diluted (5ul in 2ml LB+antibiotic) for | + | Cultures were diluted (5ul in 2ml LB+antibiotic) for Tecan Test: |
{| border='1' align='center' | {| border='1' align='center' | ||
| Line 140: | Line 139: | ||
|I74C5|| I84C5 ||I104C5 | |I74C5|| I84C5 ||I104C5 | ||
|- | |- | ||
| - | |I124C5|| | + | |I124C5||<partinfo>BBa_B0032</partinfo>|| A2 |
|} | |} | ||
| Line 224: | Line 223: | ||
|Ent4C5|| 27,6 ng/ul | |Ent4C5|| 27,6 ng/ul | ||
|- | |- | ||
| - | | | + | |<partinfo>BBa_B0034</partinfo> || 50,4 ng/ul |
|- | |- | ||
|linker (<partinfo>BBa_K105012</partinfo>) || 39,3 ng/ul | |linker (<partinfo>BBa_K105012</partinfo>) || 39,3 ng/ul | ||
|- | |- | ||
| - | |plux||45,1 ng/ul | + | |plux (<partinfo>BBa_R0062</partinfo>)||45,1 ng/ul |
|} | |} | ||
| Line 320: | Line 319: | ||
| PhaP S-S || Insert || 30+10° || 26 || 0 +7 || 1+1 XbaI || 1+1 SpeI || 3+1 | | PhaP S-S || Insert || 30+10° || 26 || 0 +7 || 1+1 XbaI || 1+1 SpeI || 3+1 | ||
|- | |- | ||
| - | | | + | | <partinfo>pSB1AK3</partinfo> || Insert || 25 || 14 || 6,5 || 1XbaI || 1 SpeI || 2,5 |
|} | |} | ||
| Line 329: | Line 328: | ||
<table align='center'><tr><td rowspan='2'> | <table align='center'><tr><td rowspan='2'> | ||
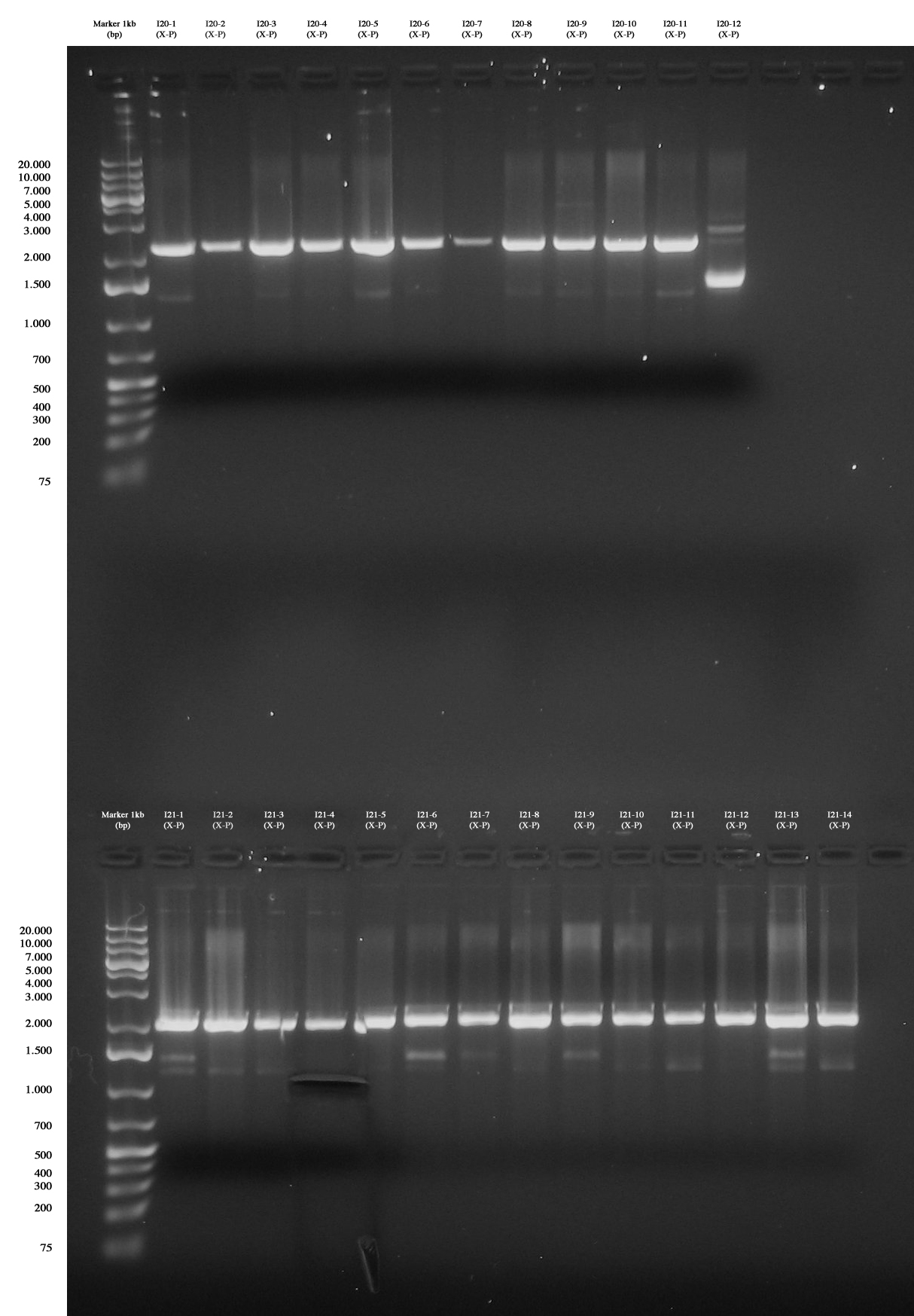
| - | [[Image:Gel_grande_10_agosto.jpg|thumb| | + | [[Image:Gel_grande_10_agosto.jpg|thumb|350px|center|Up: Screening for I20-1..12 (PhaP 10-S (X-P)): all colonies are negative :( Down: Screening for I21-1..14 (PhaP S-S (X-P)): all colonies are negative, except for I21-4 gel run/cut and purified (we ran gel again after the cut).]] |
</td><td> | </td><td> | ||
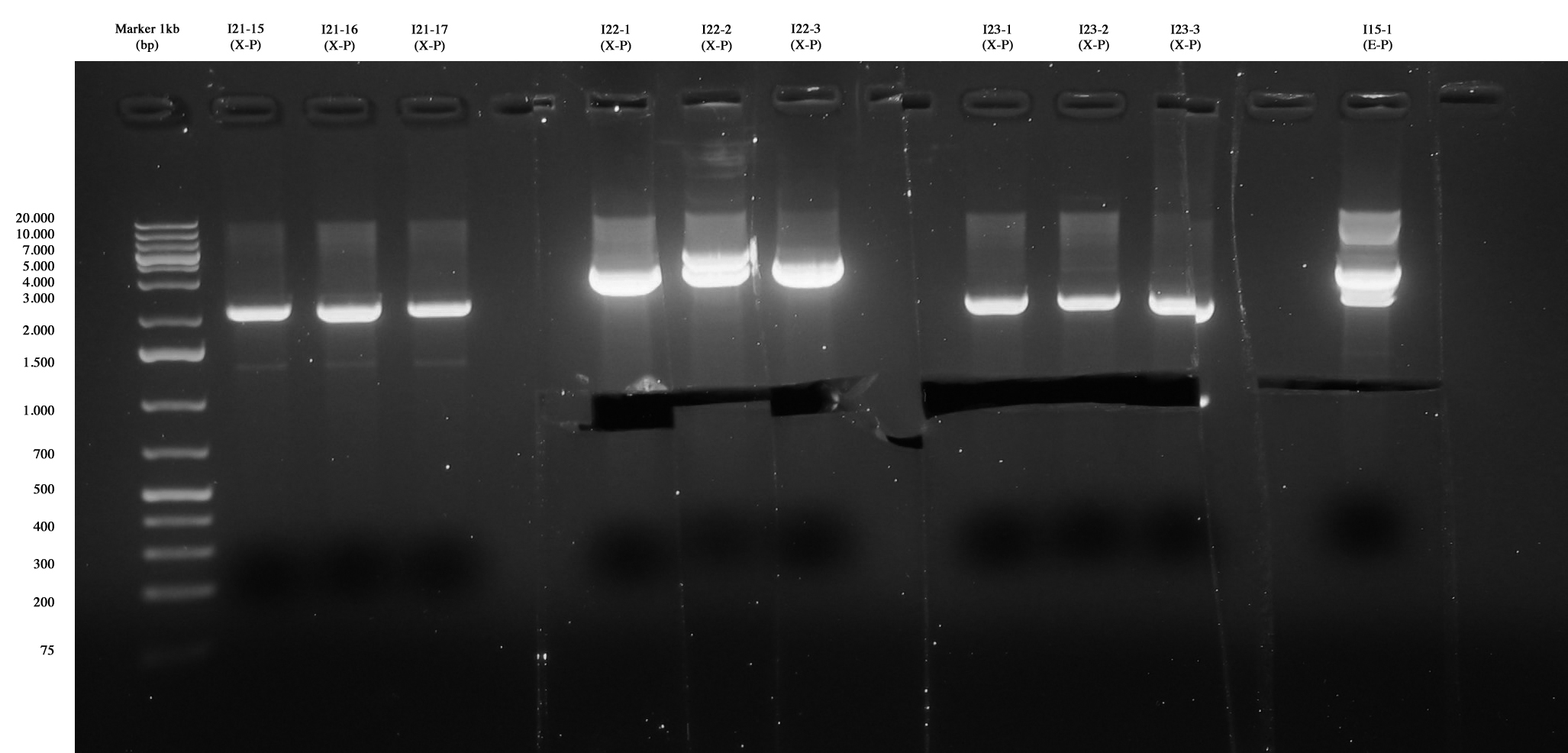
| - | [[Image:Gel_medio_1_10_agosto.jpg|thumb| | + | [[Image:Gel_medio_1_10_agosto.jpg|thumb|250px|center|Screening for I21-15..17, I22, I23 and I15-1. The three colonies of I21 were negative, I22 and I23 were positive, so they were excided and purified. I15-1 was excided, even if extra-bands were observed. We use it for ligation because sequencing is ok!]] |
</td></tr> | </td></tr> | ||
<tr> | <tr> | ||
<td> | <td> | ||
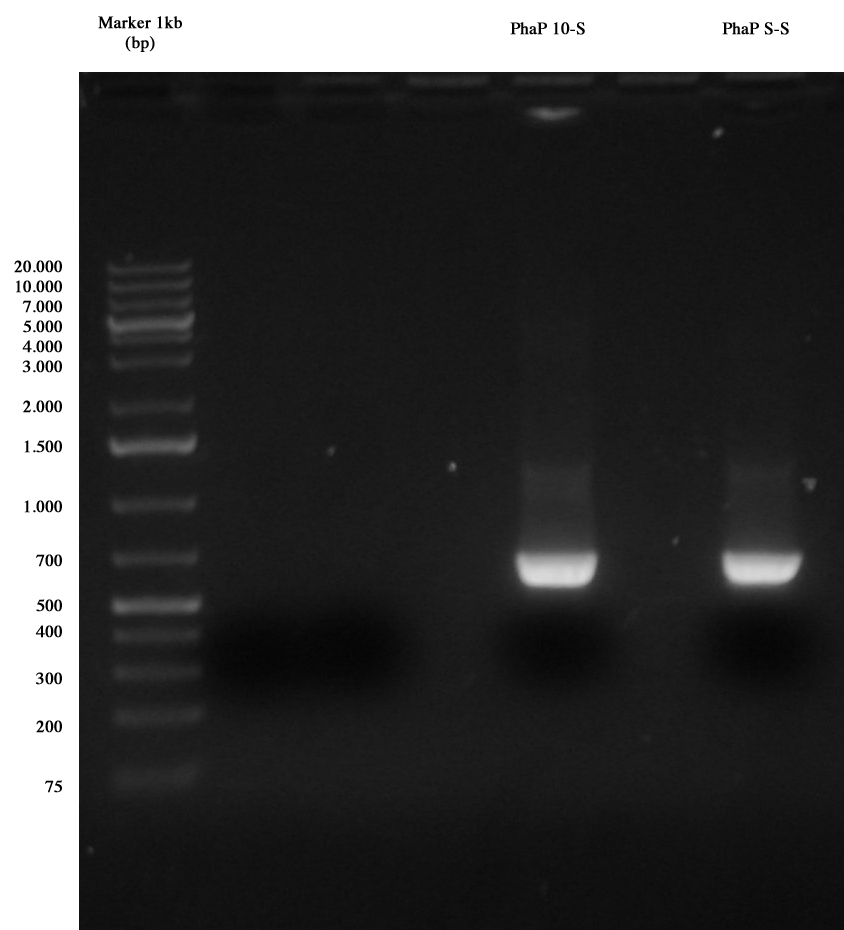
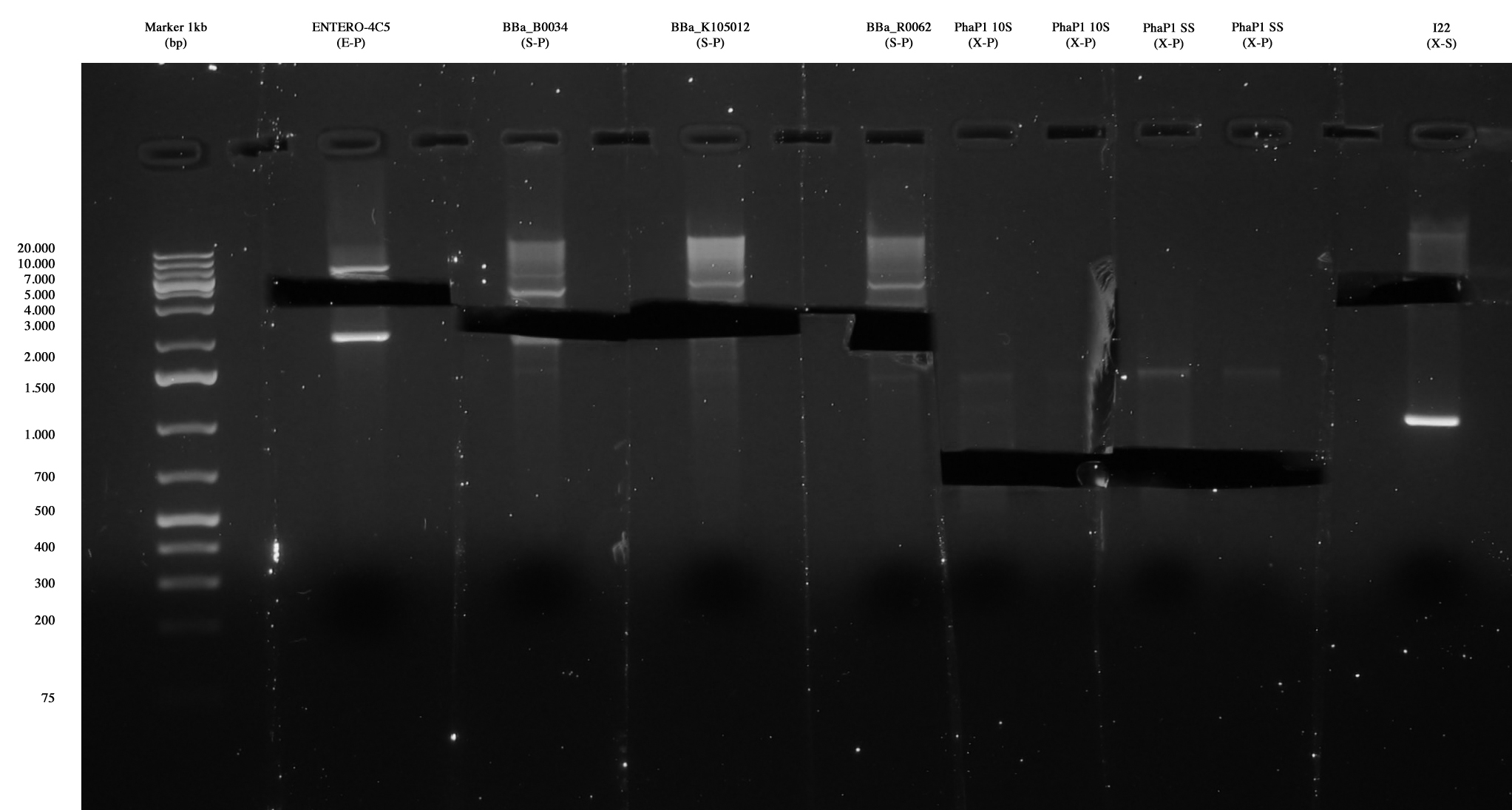
| - | [[Image:Gel_medio_2_10_agosto.jpg|thumb| | + | [[Image:Gel_medio_2_10_agosto.jpg|thumb|250px|center|Ent4C5, <partinfo>BBa_B0034</partinfo>, <partinfo>BBa_K105012</partinfo> and <partinfo>BBa_R0062</partinfo> were extracted and purified. Also PhaP 10-S (X-S) and PhaP S-S (X-S) were extracted and purified, in order to repeat the ligation with a vector, since one only colony was positive for I21 and no colony was positive for I20. <partinfo>pSB1AK3</partinfo> vector cut X-S was also gel extracted and purified.]] |
</td></tr></table> | </td></tr></table> | ||
After purification, digested DNA was quantified as follows: | After purification, digested DNA was quantified as follows: | ||
| - | {| border= | + | {| border="1" align="center" |
| - | | I21-4 (X-P)|| ng/ul | + | | I21-4 (X-P)|| 1,7 ng/ul |
|- | |- | ||
| - | | I22-1 (X-P)|| ng/ul | + | | I22-1 (X-P)|| 11,7 ng/ul |
|- | |- | ||
| - | | I22-2 (X-P) || ng/ul | + | | I22-2 (X-P) || 6,1 ng/ul |
|- | |- | ||
| - | | I22-3 (X-P) || ng/ul | + | | I22-3 (X-P) || 11,3 ng/ul |
|- | |- | ||
| - | | I23-1 (X-P) || ng/ul | + | | I23-1 (X-P) || 4,5 ng/ul |
|- | |- | ||
| - | | I23-2 (X-P) || ng/ul | + | | I23-2 (X-P) || 3,1 ng/ul |
|- | |- | ||
| - | | I23-3 (X-P) || ng/ul | + | | I23-3 (X-P) || 5,6 ng/ul |
|- | |- | ||
| - | | I15-1 (E-P) || ng/ul | + | | I15-1 (E-P) ||5,2 ng/ul |
|- | |- | ||
| - | | pSB4C5 (E-P) || ng/ul | + | | <partinfo>pSB4C5</partinfo> (E-P) ||6,0 ng/ul |
|- | |- | ||
| - | | <partinfo>BBa_B0034</partinfo> (E-P) || ng/ul | + | | <partinfo>BBa_B0034</partinfo> (E-P) ||20,9 ng/ul |
|- | |- | ||
| - | | linker ( | + | | linker (<partinfo>BBa_K105012</partinfo>) <br/> (S-P) ||17,6 ng/ul |
|- | |- | ||
| - | |pLux (S-P) || ng/ul | + | |pLux (<partinfo>BBa_R0062</partinfo>) <br/> (S-P) ||16,8 ng/ul |
|- | |- | ||
| - | | PhaP 10-S (X-S) || ng/ul | + | | PhaP 10-S (X-S) || 14,0 ng/ul |
|- | |- | ||
| - | | PhaP S-S (X-S) || ng/ul | + | | PhaP S-S (X-S) ||13,0 ng/ul |
|- | |- | ||
| - | | | + | | <partinfo>pSB1AK3</partinfo> (X-S) ||18,6 ng/ul |
|} | |} | ||
| + | |||
| + | |||
| + | Ligation of: | ||
| + | |||
| + | * I15_4C5= I15-1 (E-P)+pSB4C5(E-P) | ||
| + | * I24=I22-1(X-P)+Plux(S-P) | ||
| + | * I26=linker(S-P)+I21-4(X-P) | ||
| + | * I20_M=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) not dephosphorylized | ||
| + | * I21_M=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) not dephosphorylized | ||
| + | * I20_A=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylized with antarctic phosphatase | ||
| + | * I21_A=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylized with antarctic phosphatase | ||
| + | * I20_C=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylized with alkaline phosphatase (from calf intestine) | ||
| + | * I21_C=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylized with alkaline phosphatase (from calf intestine) | ||
| + | |||
| + | Ligations were incubated ON at 16°C and tomorrow will be transformed. | ||
| + | |||
| + | ---- | ||
| + | Colonies transformed with RING, F2620-4C5, Nothing and plated on proper agar plates were still too little to check efficiency of MC123/008 and MG123/008 competent cells. So we let them grow another day and night at 30°C. | ||
| + | |||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 11th== | ==August, 11th== | ||
| + | |||
| + | Trasformation of ligations in: | ||
| + | <table width='90%' border='1'> | ||
| + | <tr><th>Ligation name</th><th>E. coli strain </th><th> Resistance </th></tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I154C5=I15-1 (E-P) + pSB4C5 (E-P) | ||
| + | </td> | ||
| + | <td> | ||
| + | TOP10 | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr><tr><td> | ||
| + | * I24=I22-1(X-P)+Plux(S-P)</td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | |||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | |||
| + | * I26=linker(S-P)+I21-4(X-P) | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | |||
| + | </td> | ||
| + | <td>Amp 100 </td> | ||
| + | </tr><tr><td> | ||
| + | * I20_M=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) not dephosphorylated | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | * I21_M=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) not dephosphorylated | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | * I20_A=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylated with antarctic phosphatase | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | * I21_A=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylated with antarctic phosphatase | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | * I20_C=PhaP 10-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylated with alkaline phosphatase (from calf intestine) | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr><tr><td> | ||
| + | * I21_C=PhaP S-S (X-S)+<partinfo>pSB1AK3</partinfo> (X-S) dephosphorylated with alkaline phosphatase (from calf intestine) | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Amp 100</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | Plates were incubated overnight at 37°C. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
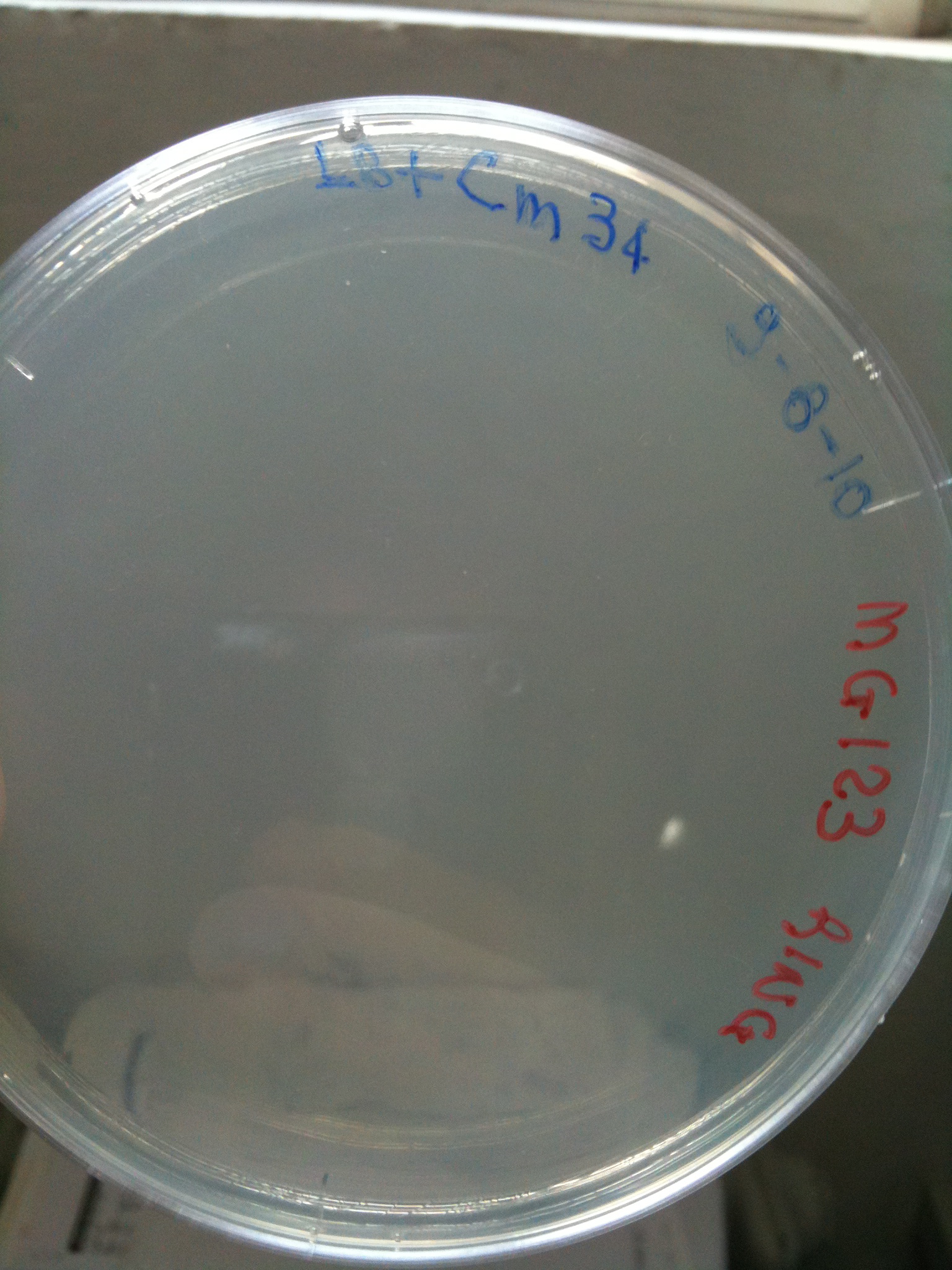
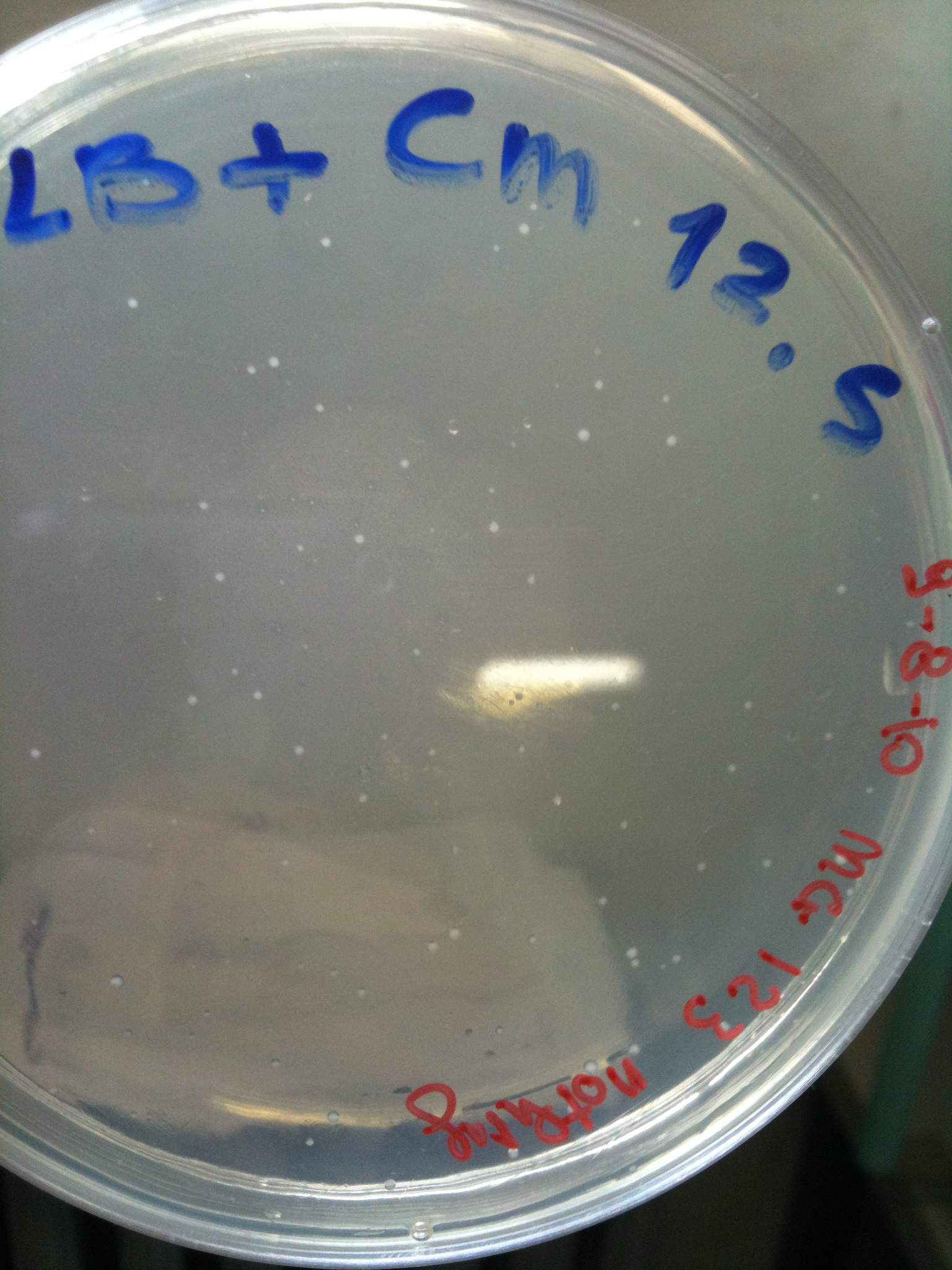
| + | Plates grown at 30°C showed the following results: | ||
| + | {|align="center" border="1" | ||
| + | | ||'''MC123'''||'''MC008'''||'''MG123'''||'''MG008''' | ||
| + | |- | ||
| + | |'''F2620-4C5'''||Grown (OK)||Grown (OK)||Grown (OK - but only ~10 colonies)||Grown (OK) | ||
| + | |- | ||
| + | |'''RING'''|| Not grown (OK)||Not plated||Not grown (OK)||Grown (OK) | ||
| + | |- | ||
| + | |'''NOTHING'''||Not grown (OK)||Not grown (OK)||Grown (???)||Not grown (OK) | ||
| + | |} | ||
| + | |||
| + | {| align="center" | ||
| + | | [[Image:UNIPV_Pavia10_MC123_F2620_4C5.jpg|thumb|200px|center|MC123 transformed with F2620-4C5 (OK)]] || [[Image:UNIPV_Pavia10_MC123_RING.jpg|thumb|200px|center|MC123 transformed with RING (OK)]] || [[Image:UNIPV_Pavia10_MC123_NOTHING.jpg|thumb|200px|center|MC123 transformed with NOTHING (OK)]] | ||
| + | |- | ||
| + | | [[Image:UNIPV_Pavia10_MC008_F2620_4C5.jpg|thumb|200px|center|MC008 transformed with F2620-4C5 (OK)]] || MC008 transformed with RING<br/>(Not plated) || [[Image:UNIPV_Pavia10_MC008_NOTHING.jpg|thumb|200px|center|MC008 transformed with NOTHING (OK)]] | ||
| + | |- | ||
| + | | [[Image:UNIPV_Pavia10_MG123_F2620_4C5.jpg|thumb|200px|center|MG123 transformed with F2620-4C5 (OK - but only ~10 colonies)]] || [[Image:UNIPV_Pavia10_MG123_RING.jpg|thumb|200px|center|MG123 transformed with RING (OK)]] || [[Image:UNIPV_Pavia10_MG123_NOTHING.jpg|thumb|200px|center|MG123 transformed with NOTHING (???)]] | ||
| + | |- | ||
| + | | [[Image:UNIPV_Pavia10_MG008_F2620_4C5.jpg|thumb|200px|center|MG008 transformed with F2620-4C5 (OK)]] || [[Image:UNIPV_Pavia10_MG008_RING.jpg|thumb|200px|center|MG008 transformed with RING (OK)]] || [[Image:UNIPV_Pavia10_MG008_NOTHING.jpg|thumb|200px|center|MG008 transformed with NOTHING (OK)]] | ||
| + | |} | ||
| + | |||
| + | So we decided to store plates at +4°C (in order to calculate efficiency in the next days) and to repeat transformation for | ||
| + | *MC008: F2620-4C5 (Cm 12,5 ug/ml), RING (Cm 34 ug/ml) | ||
| + | *MG123: F2620-4C5 (Cm 12,5 ug/ml), RING (Cm 34 ug/ml), NOTHING (12,5 ug/ml) | ||
| + | We transformed 1 ul (~4 ng of DNA) and plates were let grow at 30°C for about twenty hours. | ||
| + | |||
| + | ---- | ||
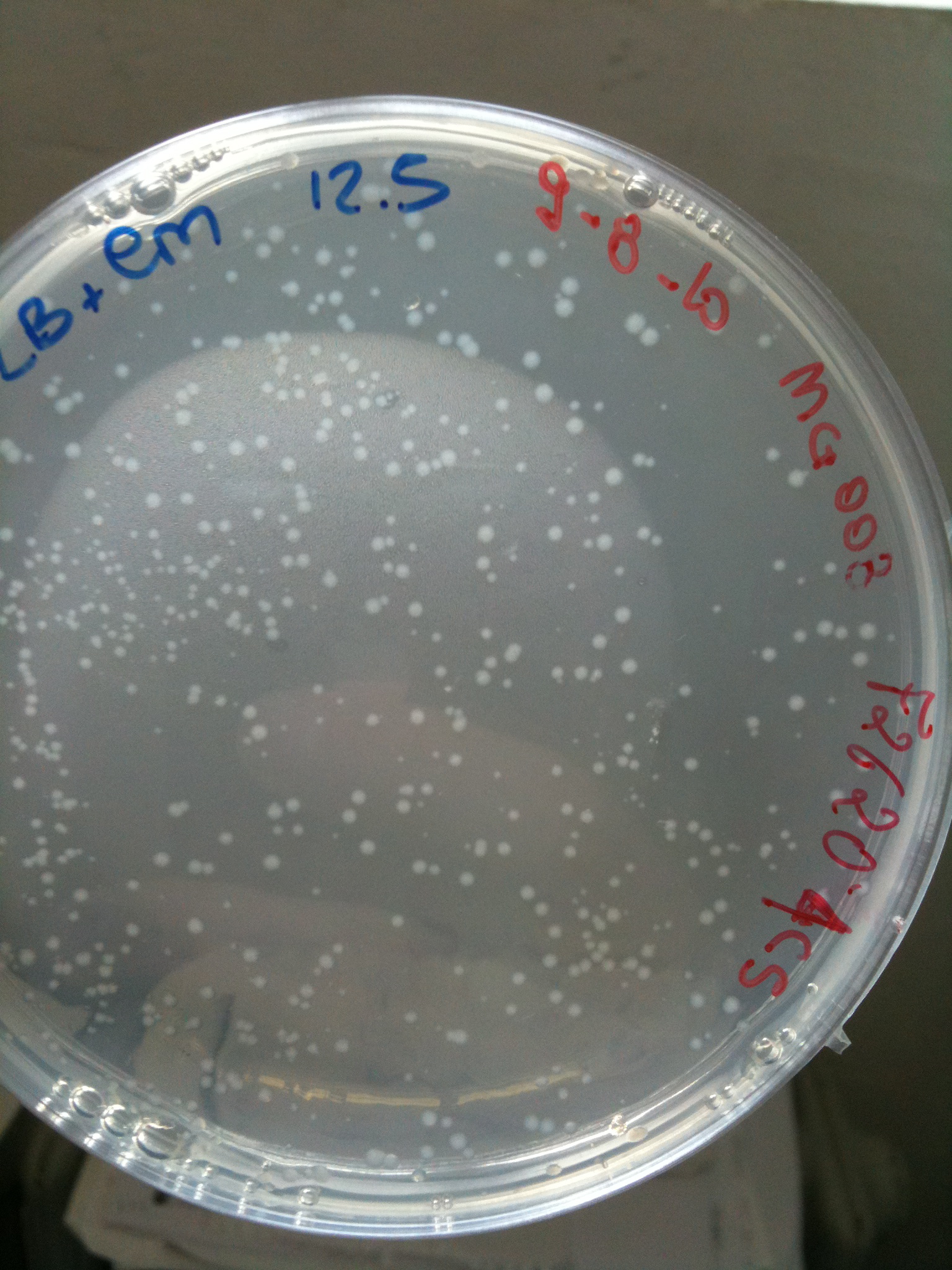
| + | Results for our test with Cm 6 ug/ml agar plates: | ||
| + | [[Image:UNIPV_Pavia10_Cm6PlateTest.jpg|thumb|200px|center|Test-plate Cm 6 ug/ml: F2620-4C5 is the positive control]] | ||
| + | |||
| + | As you can see not only positive control F2620-4C5 grew but also very small colonies for MG1655, MG123, MC008. | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 12th== | ==August, 12th== | ||
| + | |||
| + | This morning we checked if our plates were grown. | ||
| + | I24 showed many colonies, just like I26. | ||
| + | I15_4C5 showed 3 colonies. | ||
| + | I20_M and I21_M showed many colonies | ||
| + | |||
| + | 3 colonies of I24 were picked and incubated in 1 ml LB+Amp. After 6hours glycerol stock was prepared and falcon tubes were re-filled with 5ml LB+Amp in order to perform a screening tomorrow. All 3 colonies of I15_4C5 were picked and incubated in 1 ml LB+Cm 12,5. They were stocked, too, and refilled for further screening. | ||
| + | |||
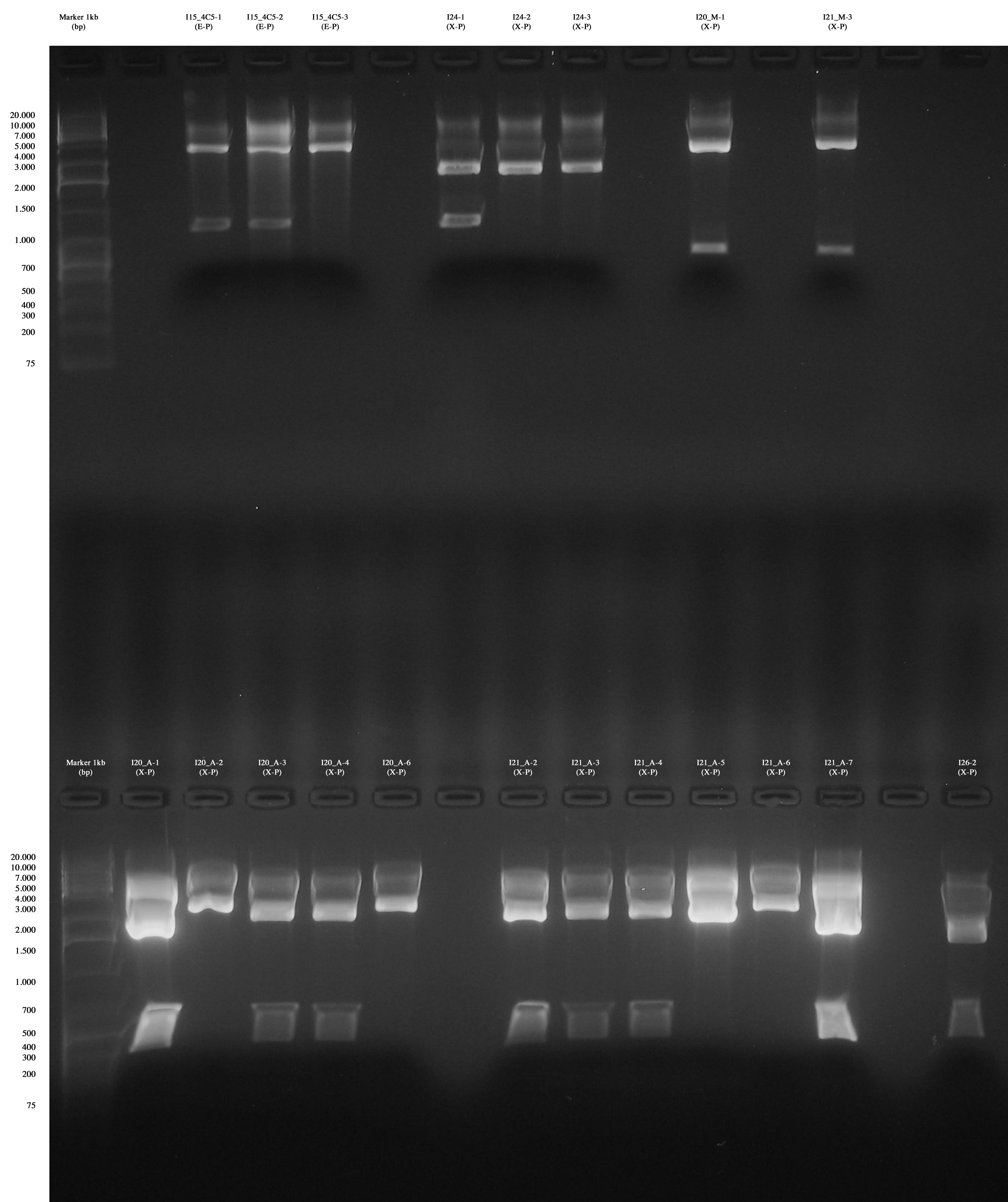
| + | Today we decided to perform a massive screening for phasins. For this reason, we decided to perform a colony PCR for: | ||
| + | |||
| + | |||
| + | <table border='2' align='center'><tr><th rowspan='3'>Culture<br> name</th> | ||
| + | <td>I20_M-1</td> | ||
| + | <td>I21_M-2</td> | ||
| + | <td>I20_M-3</td> | ||
| + | <td>I20_M-4</td> | ||
| + | <td>I21_M-1</td> | ||
| + | <td>I21_M-2</td> | ||
| + | <td>I21_M-3</td> | ||
| + | <td>I21_M-4</td> | ||
| + | <td>I20_A-1</td> | ||
| + | <td>I20_A-2</td> | ||
| + | <td>I20_A-3</td> | ||
| + | <td>I20_A-4</td></tr> | ||
| + | <tr> | ||
| + | |||
| + | <td>I20_A-5</td> | ||
| + | <td>I21_A-6</td> | ||
| + | <td>I20_A-7</td> | ||
| + | <td>I21_A-1</td> | ||
| + | <td>I21_A-2</td> | ||
| + | <td>I21_A-3</td> | ||
| + | <td>I21_A-4</td> | ||
| + | <td>I21_A-5</td> | ||
| + | <td>I21_A-6</td> | ||
| + | <td>I21_A-7</td> | ||
| + | <td>I20_C-1</td> | ||
| + | <td>I20_C-2</td></tr> | ||
| + | <tr> | ||
| + | |||
| + | <td>I20_C-3</td> | ||
| + | <td>I20_C-4</td> | ||
| + | <td>I21_C-1</td> | ||
| + | <td>I21_C-2</td> | ||
| + | <td>I21_C-3</td> | ||
| + | <td>I21_C-4</td> | ||
| + | <td>I21_C-5</td> | ||
| + | <td>I26-1</td> | ||
| + | <td>I26-2</td> | ||
| + | <td>I26-3</td> | ||
| + | <td>I26-4</td> | ||
| + | <td>I26-5</td></tr> | ||
| + | </table> | ||
| + | |||
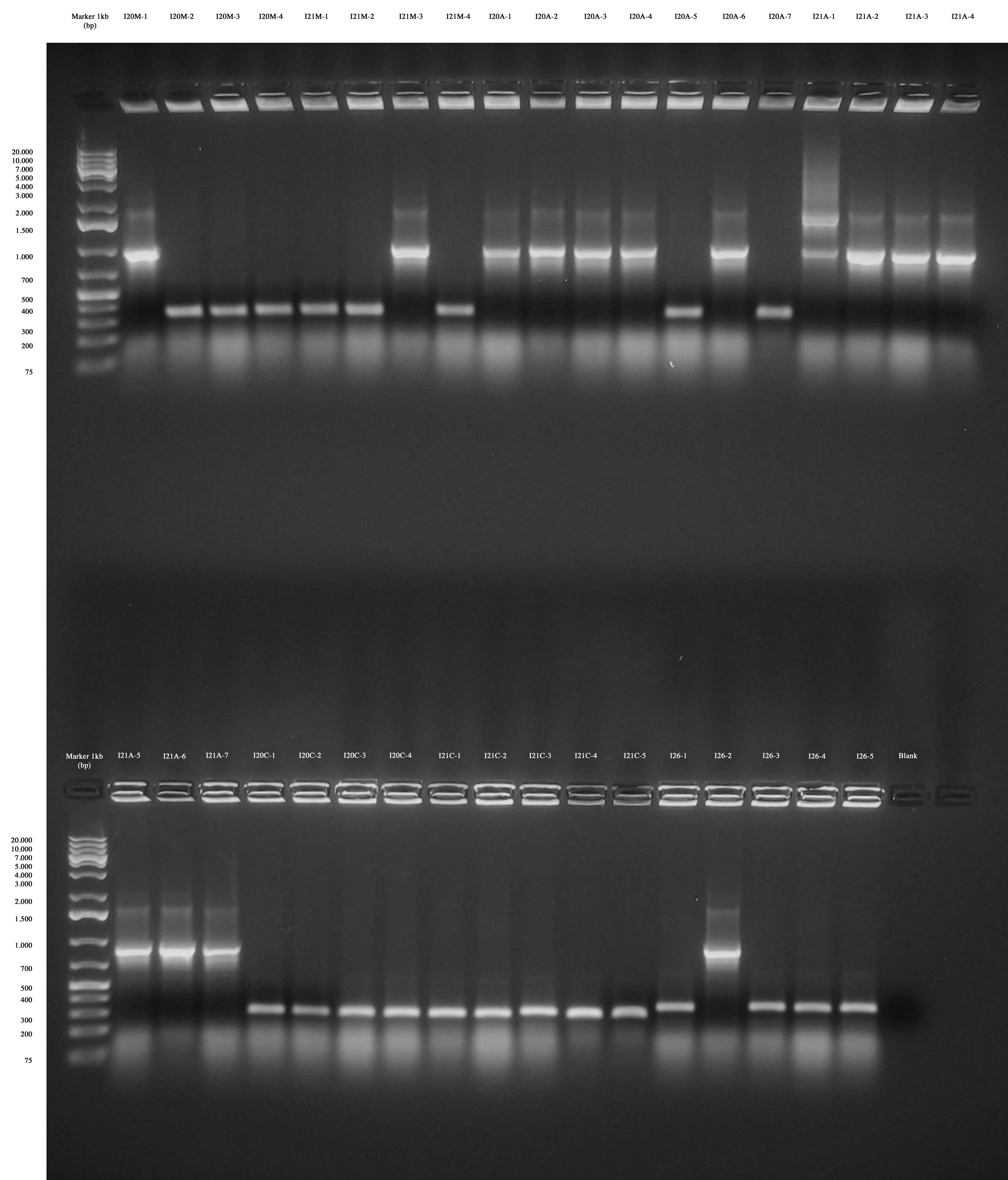
| + | [[Image:UNIPV_Pavia12_8_10_PhapScreeningPCR.jpg|thumb|600px|center|Big gel run]] | ||
| + | |||
| + | Screening showed that positive colonies bearing the insert are: | ||
| + | *I20_M-1 | ||
| + | *I21_M-3 | ||
| + | *I20_A-1 | ||
| + | *I20_A-2 | ||
| + | *I20_A-3 | ||
| + | *I20_A-4 | ||
| + | *I20_A-6 | ||
| + | *I21_A-2 | ||
| + | *I21_A-3 | ||
| + | *I21_A-4 | ||
| + | *I21_A-5 | ||
| + | *I21_A-6 | ||
| + | *I21_A-7 | ||
| + | *I26-2 | ||
| + | For these cultures an inoculum was performed in 6ml LB+Amp. Tomorrow we will prepare glycerol stocks for them and we will perform a further screening, with an X-P digestion in order to see if the insert was ligated in the right direction. | ||
| + | |||
| + | Glycerol stock was prepared for: | ||
| + | * I15_4C5-1 | ||
| + | * I15_4C5-2 | ||
| + | * I15_4C5-3 | ||
| + | * I24-1 | ||
| + | * I24-2 | ||
| + | * I24-3 | ||
| + | |||
| + | and falcon tubes were re-filled with fresh LB added with proper antibiotic. Tomorrow MiniPrep will be performed for also these cultures and plasmids will be digested and gel run. | ||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
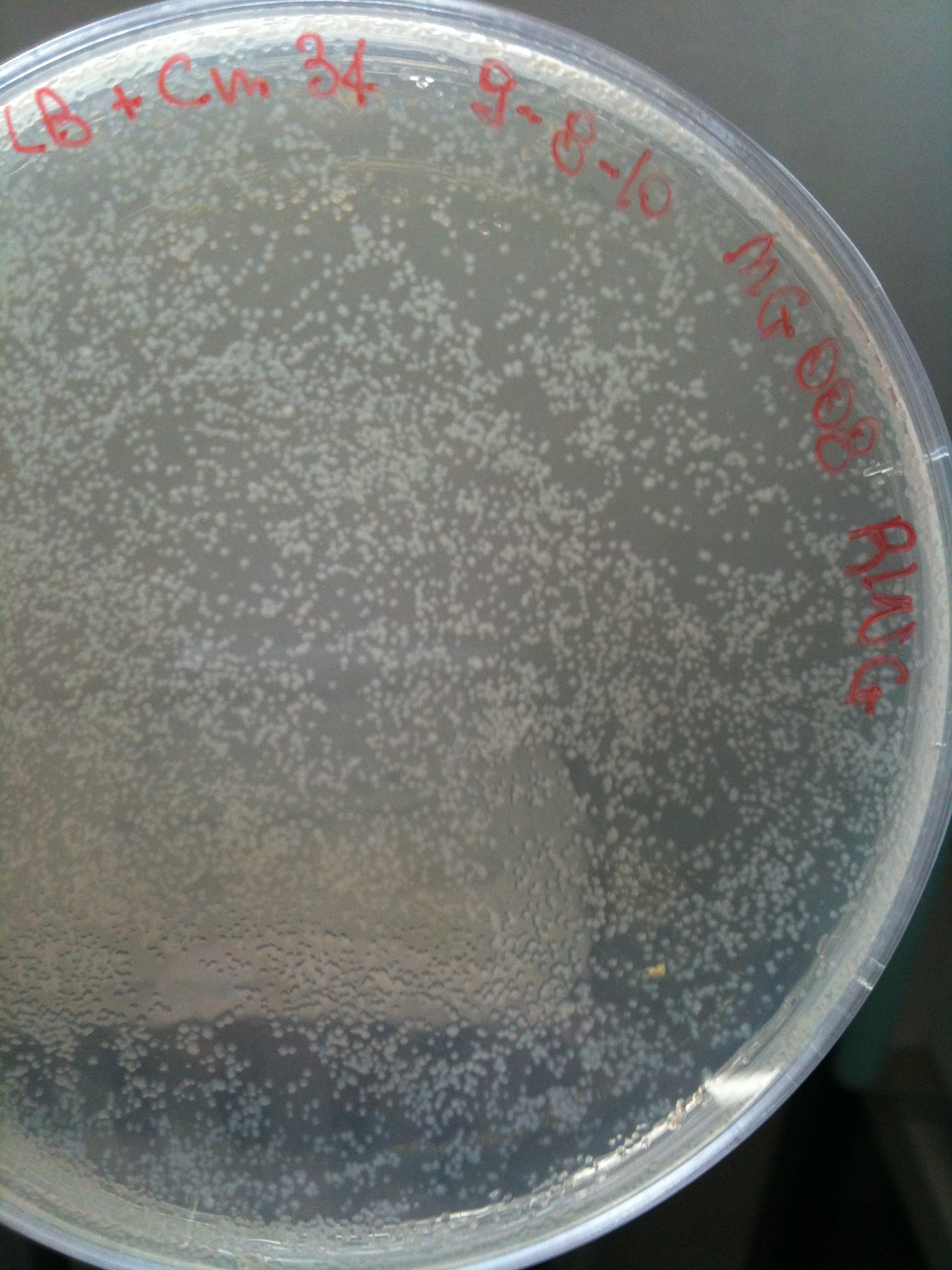
| + | Only MC008 transformed with RING was grown, so we stored it at +4°C with the other transformed plates and let the others grow at 30°C. We would have calculated the transformation efficiency for all plates together. | ||
| + | [[Image:UNIPV_Pavia10_MC008_RING2.jpg|thumb|200px|center|MC008 transformed again with RING (OK)]] | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 13th== | ==August, 13th== | ||
| + | Today Mr.Gene came to visit and he brought us two beautiful gifts: Intein and CREAM !! They were stored at +4°C. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | Here the last four plates of efficiency test: | ||
| + | {|align="center" | ||
| + | |colspan="3"|[[Image:UNIPV_Pavia10_MC008_F2620_4C52.jpg|thumb|200px|center|MC008 transformed again with F2620-4C5 (OK, but badly plated)]] | ||
| + | |- | ||
| + | |[[Image:UNIPV_Pavia10_ MG123_F2620_4C52.jpg|thumb|200px|center| MG123 transformed again with F2620_4C5 (OK)]] || [[Image:UNIPV_Pavia10_ MG123_RING2.jpg|thumb|200px|center| MG123 transformed again with RING (OK)]] || [[Image:UNIPV_Pavia10_ MG123_NOTHING2.jpg|thumb|200px|center| MG123 transformed again with NOTHING (OK)]] | ||
| + | |} | ||
| + | |||
| + | We calculated (thanks Alessandro, Chiara, Riccardo) efficiency of our home-made competent cells MC123/008 and MG123/008 (#colonies/ug DNA): | ||
| + | {|align="center" border="1" | ||
| + | |colspan="2"|'''Culture'''||'''#colonies'''||'''Efficiency''' | ||
| + | |- | ||
| + | |rowspan="3"|'''MC123'''||''F2020-4C5''||218||~10^4 | ||
| + | |- | ||
| + | |''RING''||0 (OK)||- | ||
| + | |- | ||
| + | |''NOTHING''||0 (OK)||- | ||
| + | |- | ||
| + | |rowspan="3"|'''MC008'''||''F2020-4C5''||colspan="2"|OK but badly plated: not countable | ||
| + | |- | ||
| + | |''RING''||814||~10^5 | ||
| + | |- | ||
| + | |''NOTHING''||0 (OK)||- | ||
| + | |- | ||
| + | |rowspan="3"|'''MG123'''||''F2020-4C5''||42||~10^4 | ||
| + | |- | ||
| + | |''RING''||0 (OK)||- | ||
| + | |- | ||
| + | |''NOTHING''||0 (OK)||- | ||
| + | |- | ||
| + | |rowspan="3"|'''MG008'''||''F2020-4C5''||693||~10^5 | ||
| + | |- | ||
| + | |''RING''||7944||~10^6 | ||
| + | |- | ||
| + | |''NOTHING''||0 (OK)||- | ||
| + | |} | ||
| + | |||
| + | ---- | ||
| + | |||
| + | Glycerol stock was prepared for: | ||
| + | * I20_M-1 | ||
| + | * I21_M-3 | ||
| + | * I20_A-1 | ||
| + | * I20_A-2 | ||
| + | * I20_A-3 | ||
| + | * I20_A-4 | ||
| + | * I20_A-6 | ||
| + | * I21_A-2 | ||
| + | * I21_A-3 | ||
| + | * I21_A-4 | ||
| + | * I21_A-5 | ||
| + | * I21_A-6 | ||
| + | * I21_A-7 | ||
| + | * I26-2 | ||
| + | |||
| + | MiniPrep was performed for the following cultures and with the following quantifications: | ||
| + | {| border='1' | ||
| + | | * I15_4C5-1 ||39,7 ng/ul | ||
| + | |- | ||
| + | | * I15_4C5-2||58,7 ng/ul | ||
| + | |- | ||
| + | | * I15_4C5-3||53,4 ng/ul | ||
| + | |- | ||
| + | | * I24-1||61,3 ng/ul | ||
| + | |- | ||
| + | |* I24-2||57,5 ng/ul | ||
| + | |- | ||
| + | | * I24-3 ||52,1 ng/ul | ||
| + | |- | ||
| + | | * I20_M-1||105,1 ng/ul | ||
| + | |- | ||
| + | | * I21_M-3||80,5 ng/ul | ||
| + | |- | ||
| + | | * I20_A-1||494,1 ng/ul | ||
| + | |- | ||
| + | |* I20_A-2||101,6 ng/ul | ||
| + | |- | ||
| + | | * I20_A-3||94,1 ng/ul | ||
| + | |- | ||
| + | | * I20_A-4||85,5 ng/ul | ||
| + | |- | ||
| + | | * I20_A-6||71,1 ng/ul | ||
| + | |- | ||
| + | | * I21_A-2||148,7 ng/ul | ||
| + | |- | ||
| + | | * I21_A-3||75,4 ng/ul | ||
| + | |- | ||
| + | | * I21_A-4||84,6 ng/ul | ||
| + | |- | ||
| + | | * I21_A-5||390,7 ng/ul | ||
| + | |- | ||
| + | | * I21_A-6||168,5 ng/ul | ||
| + | |- | ||
| + | |* I21_A-7||525,0 ng/ul | ||
| + | |- | ||
| + | | * I26-2 ||57,9 ng/ul | ||
| + | |} | ||
| + | |||
| + | Digestion was performed for: | ||
| + | {| border="1" | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer H'' | ||
| + | |- | ||
| + | | I15_4C5-1 || Screening || 25 || 4 || 17,5 || 0,5 EcoRI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I15_4C5-2 || Screening || 25 ||4 || 17,5 || 0,5 EcoRI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I15_4C5-3 || Screening || 25 ||4 || 17,5 || 0,5 EcoRI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I24-1 || Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I24-2 || Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I24-3 || Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_M-1 || Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_M-3 || Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_A-1|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_A-2|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_A-3|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_A-4|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I20_A-6|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-2|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-3|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-4|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-5|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-6|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I21_A-7|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I26-2|| Screening || 25 ||2 || 19,5 || 0,5 XbaI || 0,5 PstI || 2,5 | ||
| + | |} | ||
| + | |||
| + | |||
| + | Cultures were digested and gel run. | ||
| + | |||
| + | [[Image:UNIPV_Pavia13_agosto_2010_gel_grande.jpg|thumb|500px|center|Gel run for cultures screening]] | ||
| - | + | These are the right colonies: | |
| + | *I15-4C5-1 | ||
| + | *I15-4C5-2 | ||
| + | *I24-1 | ||
| + | *I20M-1 | ||
| + | *I21M-3 | ||
| + | *I20A-1 | ||
| + | *I20A-3 | ||
| + | *I20A-4 | ||
| + | *I21A-2 | ||
| + | *I21A-3 | ||
| + | *I21A-4 | ||
| + | *I21A-7 | ||
| + | *I26-2 | ||
| - | = | + | <div align="right"><small>[[#indice|^top]]</small></div> |
<!-- table previous next week --> | <!-- table previous next week --> | ||
| Line 387: | Line 769: | ||
<tr> | <tr> | ||
<td align="left">[[Team:UNIPV-Pavia/Calendar/August/settimana1| Previous week]]</td> | <td align="left">[[Team:UNIPV-Pavia/Calendar/August/settimana1| Previous week]]</td> | ||
| - | <td align="right">Next week</td> | + | <td align="right">[[Team:UNIPV-Pavia/Calendar/August/settimana3| Next week]]</td> |
</tr> | </tr> | ||
</table> | </table> | ||
Latest revision as of 16:58, 24 October 2010
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"