Team:TU Delft/Modeling/MFA/additional pathways
From 2010.igem.org
Lbergwerff (Talk | contribs) (→Pathways added to the E. coli metabolic network) |
Lbergwerff (Talk | contribs) |
||
| Line 2: | Line 2: | ||
{{Team:TU_Delft/frame_check}} | {{Team:TU_Delft/frame_check}} | ||
==Pathways added to the ''E. coli'' metabolic network== | ==Pathways added to the ''E. coli'' metabolic network== | ||
| + | |||
| + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 1''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] | ||
The ''E. coli'' network from Cell Net Analyzer contains, glycolysis, TCA cycle, pentose phosphate pathway, gluconeogenesis, anapleorotic routes, oxydative phosphorilization and biosynthesis pathways. CellNetAnalyzer can be found [http://www.mpi-magdeburg.mpg.de/projects/cna/cna.html here]. | The ''E. coli'' network from Cell Net Analyzer contains, glycolysis, TCA cycle, pentose phosphate pathway, gluconeogenesis, anapleorotic routes, oxydative phosphorilization and biosynthesis pathways. CellNetAnalyzer can be found [http://www.mpi-magdeburg.mpg.de/projects/cna/cna.html here]. | ||
| Line 7: | Line 9: | ||
There was one change made in the standard network for the NADH dehydrogenase reaction, annotated as NADHdehydro in CellNetAnalyzer. In the network of CellNetAnalyzer this reaction exports two protons, but this was changed to 4. | There was one change made in the standard network for the NADH dehydrogenase reaction, annotated as NADHdehydro in CellNetAnalyzer. In the network of CellNetAnalyzer this reaction exports two protons, but this was changed to 4. | ||
| - | + | ||
==Alkane degradation== | ==Alkane degradation== | ||
| Line 15: | Line 17: | ||
AlkB2 (EC 1.14.15.3) | AlkB2 (EC 1.14.15.3) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 2''' – Reaction taken from [http://metacyc.org/META/new-image?type=REACTION&object=ALKANE-1-MONOOXYGENASE-RXN Metacyc]]] |
Reaction for CNA; n-alkane + reduced rubredoxin + O2 -> n-alkanol + oxidized rubredoxin | Reaction for CNA; n-alkane + reduced rubredoxin + O2 -> n-alkanol + oxidized rubredoxin | ||
| Line 22: | Line 24: | ||
RubA3/RubA4 (EC 1.18.1.1) | RubA3/RubA4 (EC 1.18.1.1) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 3''' – Reaction taken from [http://biocyc.org/META/NEW-IMAGE?type=REACTION&object=RUBREDOXIN--NAD%2b-REDUCTASE-RXN Metacyc]]] |
Reaction for CNA; oxidized rubredoxin + NADH -> reduced rubredoxin | Reaction for CNA; oxidized rubredoxin + NADH -> reduced rubredoxin | ||
| Line 28: | Line 30: | ||
ADH (EC 1.1.1.1) | ADH (EC 1.1.1.1) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 4''' – Reaction taken from [http://biocyc.org/META/NEW-IMAGE?type=REACTION&object=ALCOHOL-DEHYDROG-GENERIC-RXN Metacyc]]] |
Reaction for CNA; n-alkanol -> n-aldehyde + NADH | Reaction for CNA; n-alkanol -> n-aldehyde + NADH | ||
| Line 34: | Line 36: | ||
ALDH (EC 1.2.1.3) | ALDH (EC 1.2.1.3) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 5''' – Reaction taken from [http://biocyc.org/META/NEW-IMAGE?type=REACTION&object=RXN-4142 Metacyc]]] |
Reaction for CNA; n-aldehyde -> n-fatty acid acid + NADH | Reaction for CNA; n-aldehyde -> n-fatty acid acid + NADH | ||
| Line 43: | Line 45: | ||
fatty acyl-CoA synthetase (EC 6.2.1.3) | fatty acyl-CoA synthetase (EC 6.2.1.3) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 6''' – Reaction taken from [http://biocyc.org/META/NEW-IMAGE?type=REACTION&object=ACYLCOASYN-RXN Ecocyc]]] |
Reaction for CNA; n-fatty acid + ATP -> n-saturated fatty acyl-CoA + AMP | Reaction for CNA; n-fatty acid + ATP -> n-saturated fatty acyl-CoA + AMP | ||
| Line 49: | Line 51: | ||
Adenylate kinase (EC 2.7.4.3) | Adenylate kinase (EC 2.7.4.3) | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 7''' – Reaction taken from [http://biocyc.org/META/NEW-IMAGE?type=REACTION&object=ADENYL-KIN-RXN Ecocyc]]] |
Reaction for CNA; ATP + AMP -> 2 ADP | Reaction for CNA; ATP + AMP -> 2 ADP | ||
| Line 55: | Line 57: | ||
Fatty acid beta-oxidation cycle (EC 1.3.99.3 EC 4.2.1.17 EC 1.1.1.35 EC 2.3.1.16)] | Fatty acid beta-oxidation cycle (EC 1.3.99.3 EC 4.2.1.17 EC 1.1.1.35 EC 2.3.1.16)] | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 8''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=FAO-PWY&detail-level=2 Ecocyc]]] |
Reaction for CNA; n-saturated fatty acyl-CoA -> (n - 2)-saturated fatty acyl-CoA + acetyl-CoA + FADH2 + NADH | Reaction for CNA; n-saturated fatty acyl-CoA -> (n - 2)-saturated fatty acyl-CoA + acetyl-CoA + FADH2 + NADH | ||
| Line 67: | Line 69: | ||
In oily environments oxygen diffuses more difficult into the water phase. The oxygen is used for the oxidative phosphorylation, regenerating NADH, and for the first step in the hydrocarbon degradation. To be more efficient with oxygen an additional electron acceptor was introduced. | In oily environments oxygen diffuses more difficult into the water phase. The oxygen is used for the oxidative phosphorylation, regenerating NADH, and for the first step in the hydrocarbon degradation. To be more efficient with oxygen an additional electron acceptor was introduced. | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 9''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] |
[http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=PWY0-1335 The standard oxydative phosphorylation (EC 1.6.5.3 EC 1.10.2.-)] | [http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=PWY0-1335 The standard oxydative phosphorylation (EC 1.6.5.3 EC 1.10.2.-)] | ||
| Line 73: | Line 75: | ||
The second step will be disabled in the network and be replaced with a nitrate reductase [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=RXN0-6369 (EC 1.7.99.4)] | The second step will be disabled in the network and be replaced with a nitrate reductase [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=RXN0-6369 (EC 1.7.99.4)] | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 10''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] |
*note: make link open up a new window | *note: make link open up a new window | ||
| Line 89: | Line 91: | ||
*note: make link open up a new window | *note: make link open up a new window | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 11''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] |
The pathway is displayed here [http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY1-3&detail-level=2 (EC 2.3.19 EC 1.1.1.36 EC 2.3.1.-)] | The pathway is displayed here [http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY1-3&detail-level=2 (EC 2.3.19 EC 1.1.1.36 EC 2.3.1.-)] | ||
| Line 107: | Line 109: | ||
The pathway is displayed here http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-6270&detail-level=2 | The pathway is displayed here http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-6270&detail-level=2 | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 12''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] |
In this scenario the lumped isoprene production pathway was added to metabolic network; | In this scenario the lumped isoprene production pathway was added to metabolic network; | ||
| Line 124: | Line 126: | ||
The pathway is displayed here http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=FHLMULTI-RXN | The pathway is displayed here http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=FHLMULTI-RXN | ||
| - | [[Image:Team_TUDelft_NADHdehydro.png | + | [[Image:Team_TUDelft_NADHdehydro.png|thumb|250px|right|'''Figure 13''' – Reaction taken from [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION-IN-PATHWAY&object=NADH-DEHYDROG-A-RXN Ecocyc]]] |
In this scenario the hydrogen production pathway was added to metabolic network; | In this scenario the hydrogen production pathway was added to metabolic network; | ||
Revision as of 10:17, 24 October 2010
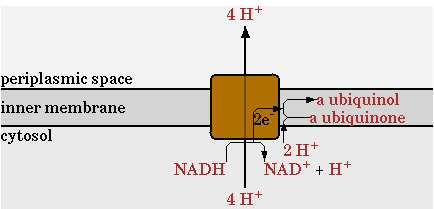
Pathways added to the E. coli metabolic network
The E. coli network from Cell Net Analyzer contains, glycolysis, TCA cycle, pentose phosphate pathway, gluconeogenesis, anapleorotic routes, oxydative phosphorilization and biosynthesis pathways. CellNetAnalyzer can be found [http://www.mpi-magdeburg.mpg.de/projects/cna/cna.html here].
There was one change made in the standard network for the NADH dehydrogenase reaction, annotated as NADHdehydro in CellNetAnalyzer. In the network of CellNetAnalyzer this reaction exports two protons, but this was changed to 4.
Alkane degradation
To link alkanes to the existing network, the beta-oxidation was chosen as entry point. Several genes were used to transform alkanes in to alkanoic acid which enters the beta oxydation cycle. These genes were:
AlkB2 (EC 1.14.15.3)
Reaction for CNA; n-alkane + reduced rubredoxin + O2 -> n-alkanol + oxidized rubredoxin
RubA3/RubA4 (EC 1.18.1.1)
Reaction for CNA; oxidized rubredoxin + NADH -> reduced rubredoxin
ADH (EC 1.1.1.1)
Reaction for CNA; n-alkanol -> n-aldehyde + NADH
ALDH (EC 1.2.1.3)
Reaction for CNA; n-aldehyde -> n-fatty acid acid + NADH
From here the genes are already present in the E. coli genome, they were not yet present in the metabolic network for E. coli in CellNetAnalyzer however.
fatty acyl-CoA synthetase (EC 6.2.1.3)
Reaction for CNA; n-fatty acid + ATP -> n-saturated fatty acyl-CoA + AMP
Adenylate kinase (EC 2.7.4.3)
Reaction for CNA; ATP + AMP -> 2 ADP
Fatty acid beta-oxidation cycle (EC 1.3.99.3 EC 4.2.1.17 EC 1.1.1.35 EC 2.3.1.16)]
Reaction for CNA; n-saturated fatty acyl-CoA -> (n - 2)-saturated fatty acyl-CoA + acetyl-CoA + FADH2 + NADH
Biomass formation
The biomass is formed by many anabolic reactions that make monomers. All the anabolic reactions start at the so called key metabolites. There are 12 key metabolites and they are all in the glycolytic pathway and the TCA cycle. In the tool they are the red metabolites.
NO3 as electron acceptor
In oily environments oxygen diffuses more difficult into the water phase. The oxygen is used for the oxidative phosphorylation, regenerating NADH, and for the first step in the hydrocarbon degradation. To be more efficient with oxygen an additional electron acceptor was introduced.
[http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=PWY0-1335 The standard oxydative phosphorylation (EC 1.6.5.3 EC 1.10.2.-)]
The second step will be disabled in the network and be replaced with a nitrate reductase [http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=RXN0-6369 (EC 1.7.99.4)]
- note: make link open up a new window
The reaction for CNA will be;
NO3- + QH2 -> NO2-
This reaction uses NO3 as an electron acceptor to regenerate NADH and export protons to generate ATP. Less protons are exported per mol of NADH, so the ATP/NADH ratio will drop compared to oxygen. The goal of implementing this pathway however, is to see how much the oxygen requirement of E. coli can be reduced.
PHB production
In previous situations the hydrocarbons were degraded only to form biomass and CO2. It is interesting to see how much product could be made from hydrocarbons. PHB is a polymer of polyhydroxybutyrate. The production pathway of PHB is well known. PHB is a solid product which is to recover in the down stream process.
- note: make link open up a new window
The pathway is displayed here [http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY1-3&detail-level=2 (EC 2.3.19 EC 1.1.1.36 EC 2.3.1.-)]
In this scenario the lumped PHB production pathway was added to metabolic network;
2 acetyl-CoA + NADPH -> (R)-3-hydroxybutanoyl-CoA
the polymerization reaction just consumes (R)-3-hydroxybutanoyl-CoA.
Isoprene production
In previous situations the hydrocarbons were degraded only to form biomass and CO2. It is interesting to see how much product could be made from hydrocarbons. Isoprene is a volatile product found in plants. E. coli will not be able to produce is in the near future, but it is an interesting product It is a very reduced product, with a similar amount of electron per carbon atom. Hydrocarbons have 6 - 6.3 electrons per carbon atom depending on the length and isoprene has 5.6 electron per carbon electron. If these values are close to each other, it has a positive influence on the maximal theoretical yield. Also the volatile nature of isoprene is very favorable for the downstream process
- note: make link open up a new window
The pathway is displayed here http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-6270&detail-level=2
In this scenario the lumped isoprene production pathway was added to metabolic network;
1 pyruvate + 1 D-glyceraldehyde-3-phosphate + 1 NADPH + 3 NADH + 3 ATP -> isoprene + CO2
isoprene export
Hydrogen production
In previous situations the hydrocarbons were degraded only to form biomass and CO2. It is interesting to see how much product could be made from hydrocarbons. Hydrogen is considered a green fuel. Hydrogen is a volatile product and is easily separated from fermentation broth. It does however contain no carbon atoms, so hydrogen will result in production of CO2 and biomass.
- note: make link open up a new window
The pathway is displayed here http://biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=FHLMULTI-RXN
In this scenario the hydrogen production pathway was added to metabolic network;
Formate + H+ -> H2 + CO2
hydrogen export
 "
"