Team:UNIPV-Pavia/Project/solution
From 2010.igem.org
| Line 41: | Line 41: | ||
<br> | <br> | ||
| - | <table align="center" border=" | + | <table align="center" border="0" width="80%"> |
<tr> | <tr> | ||
| - | <td align="center" valign = "center" style="padding:20px"> | + | <td align="center" valign = "center" style="padding:20px" width="25%"> |
| - | <html | + | <html> |
<img src="" width="75px" height="75px"/> | <img src="" width="75px" height="75px"/> | ||
| - | + | </html> | |
| - | </html | + | <br> |
| - | < | + | [[Team:UNIPV-Pavia/Project/solution #Self-inducible promoters|Self-inducible promoters]] |
| - | + | </td> | |
| - | + | <td align="center" valign = "center" style="padding:20px" width="25%"> | |
| - | + | <html> | |
| - | <td align="center" valign = "center" style="padding:20px"> | + | |
| - | <html | + | |
<img src="" width="75px" height="75px"/> | <img src="" width="75px" height="75px"/> | ||
| - | </ | + | </html> |
| - | < | + | <br> |
| - | <td align="center" valign = "center" style="padding:20px"> | + | [[Team:UNIPV-Pavia/Project/solution #Integrative standard vectors for E. coli|Integrative standard vectors for E. coli]] |
| - | <html>< | + | </td> |
| + | <td align="center" valign = "center" style="padding:20px" width="25%"> | ||
| + | <html> | ||
| + | <img src="" width="75px" height="75px"/> | ||
| + | </html> | ||
| + | <br> | ||
| + | [[Team:UNIPV-Pavia/Project/solution #Integrative standard vectors for yeast|Integrative standard vectors for yeast]] | ||
| + | </td> | ||
| + | <td align="center" valign = "center" style="padding:20px" width="25%"> | ||
| + | <html> | ||
<img src="" width="75px" height="75px"/></a> | <img src="" width="75px" height="75px"/></a> | ||
| - | </html></td> | + | </html> |
| + | <br> | ||
| + | [[Team:UNIPV-Pavia/Project/solution #Self-cleaving affinity tags to easily purify proteins|Purification of proteins]] | ||
| + | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| Line 67: | Line 77: | ||
<table align="center" border="0" width="80%"> | <table align="center" border="0" width="80%"> | ||
=Self-inducible promoters= | =Self-inducible promoters= | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
=Integrative standard vector for E. coli= | =Integrative standard vector for E. coli= | ||
| Line 194: | Line 205: | ||
''efficiency [CFU/ug of DNA]= # CFU * 1000 ng of DNA / amount of transformed DNA [ng]'' | ''efficiency [CFU/ug of DNA]= # CFU * 1000 ng of DNA / amount of transformed DNA [ng]'' | ||
</div> | </div> | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
=Integrative standard vectors for yeast= | =Integrative standard vectors for yeast= | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
=Self-cleaving affinity tags to easily purify proteins= | =Self-cleaving affinity tags to easily purify proteins= | ||
| - | + | <div align="right"><small>[[#indice|^top]]</small></div> | |
</table> | </table> | ||
Revision as of 13:25, 22 October 2010
|
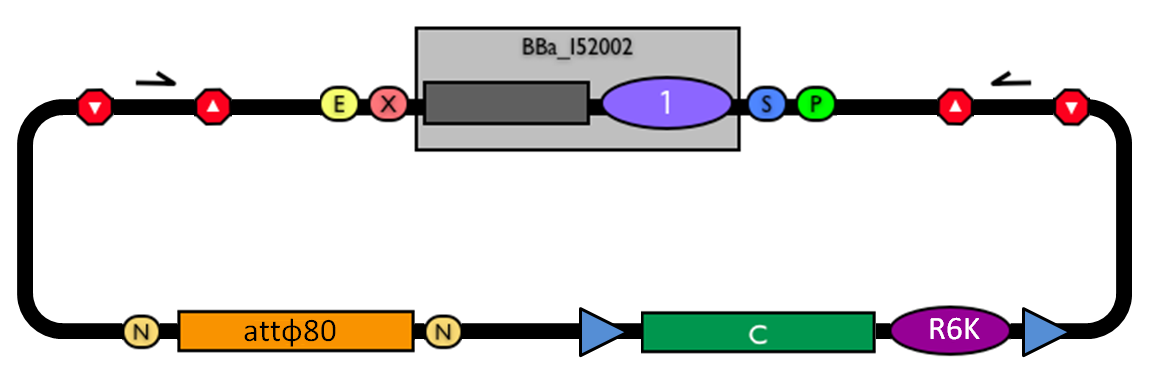
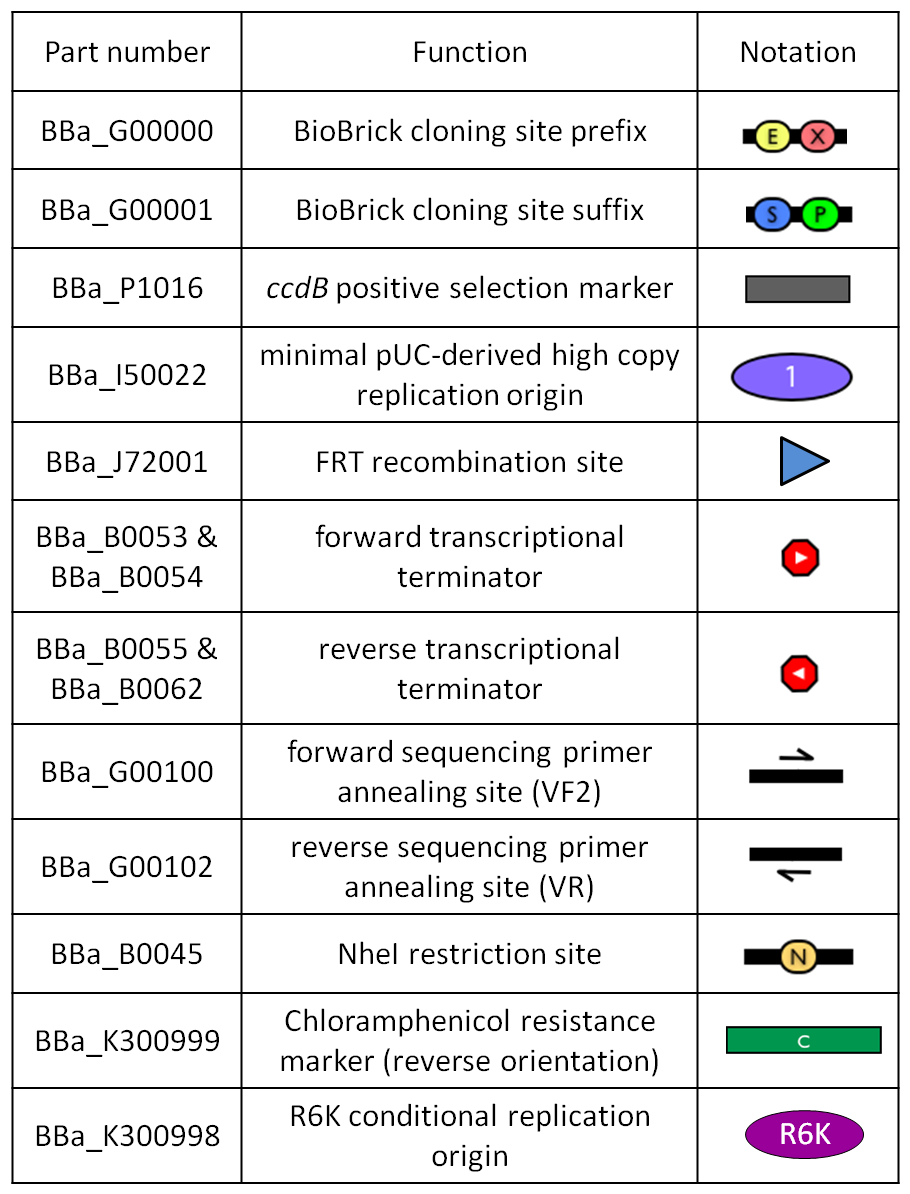
This vector can be considered as a base vector, which can be specialized to target the desired integration site in the host genome. The default version of this backbone has the bacteriophage Phi80 attP (<partinfo>BBa_K300991</partinfo>) as integration site. This vector enables multiple integrations in different positions of the same genome.
GlossaryThe passenger is the desired DNA part to be integrated into the genome. The guide is the DNA sequence that is used to target the passenger into a specific locus in the genome.
The main design features for vector engineering and for the genome integration of the vector are reported below. Vector engineering features:
Genome integration features:
How to use it<partinfo>BBa_K300000</partinfo> can be:
How to propagate it before performing genome integrationThe default version of this vector contains the <partinfo>BBa_I52002</partinfo> insert, so it *must* be propagated in a ccdB-tolerant strain such as DB3.1 (<partinfo>BBa_V1005</partinfo>). After the insertion of the desired BioBrick part in the cloning site, this vector does not contain a standard replication origin anymore, so it *must* be propagated in a pir+ or pir-116 strain such as BW25141 (<partinfo>BBa_K300984</partinfo>) or BW23474 (<partinfo>BBa_K300985</partinfo>) that can replicate the R6K conditional origin (<partinfo>BBa_J61001</partinfo>).
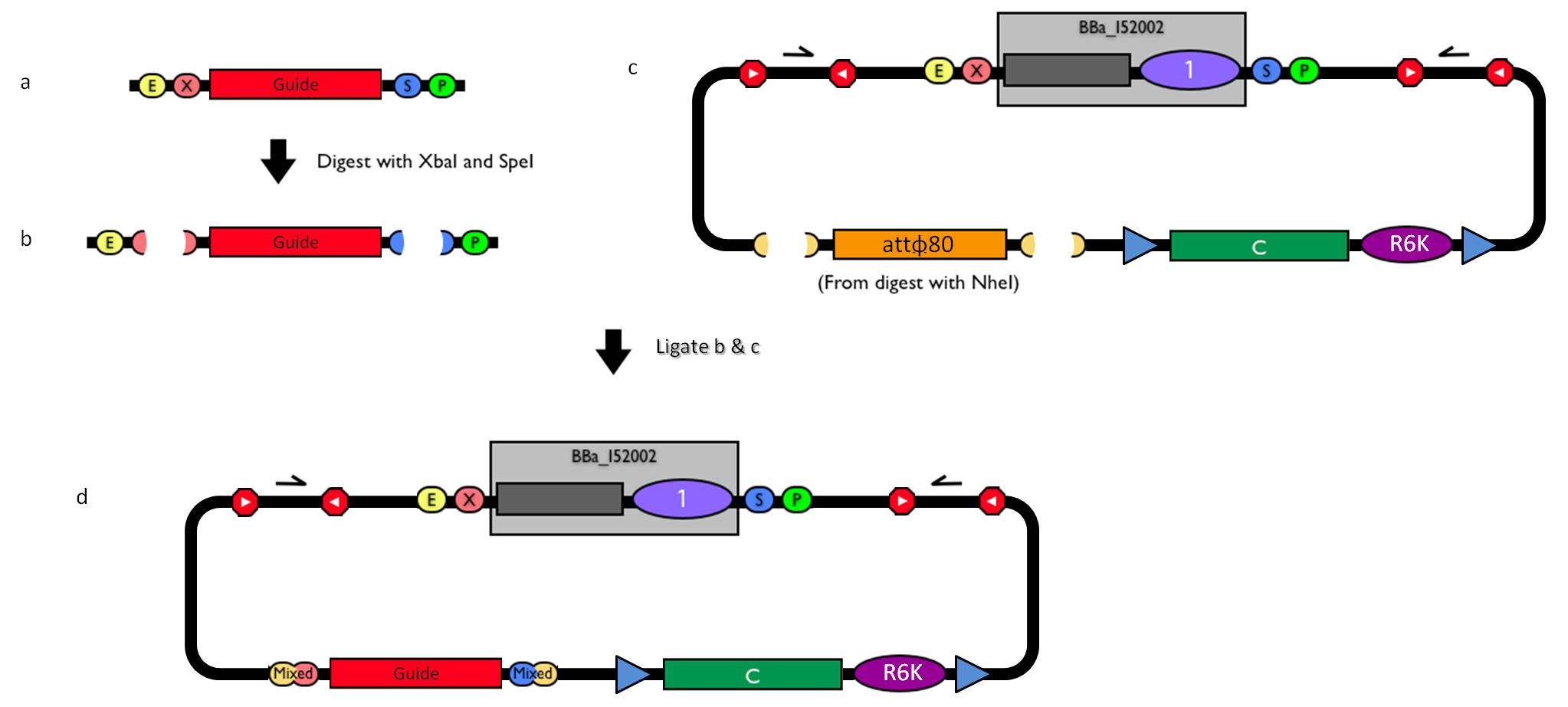
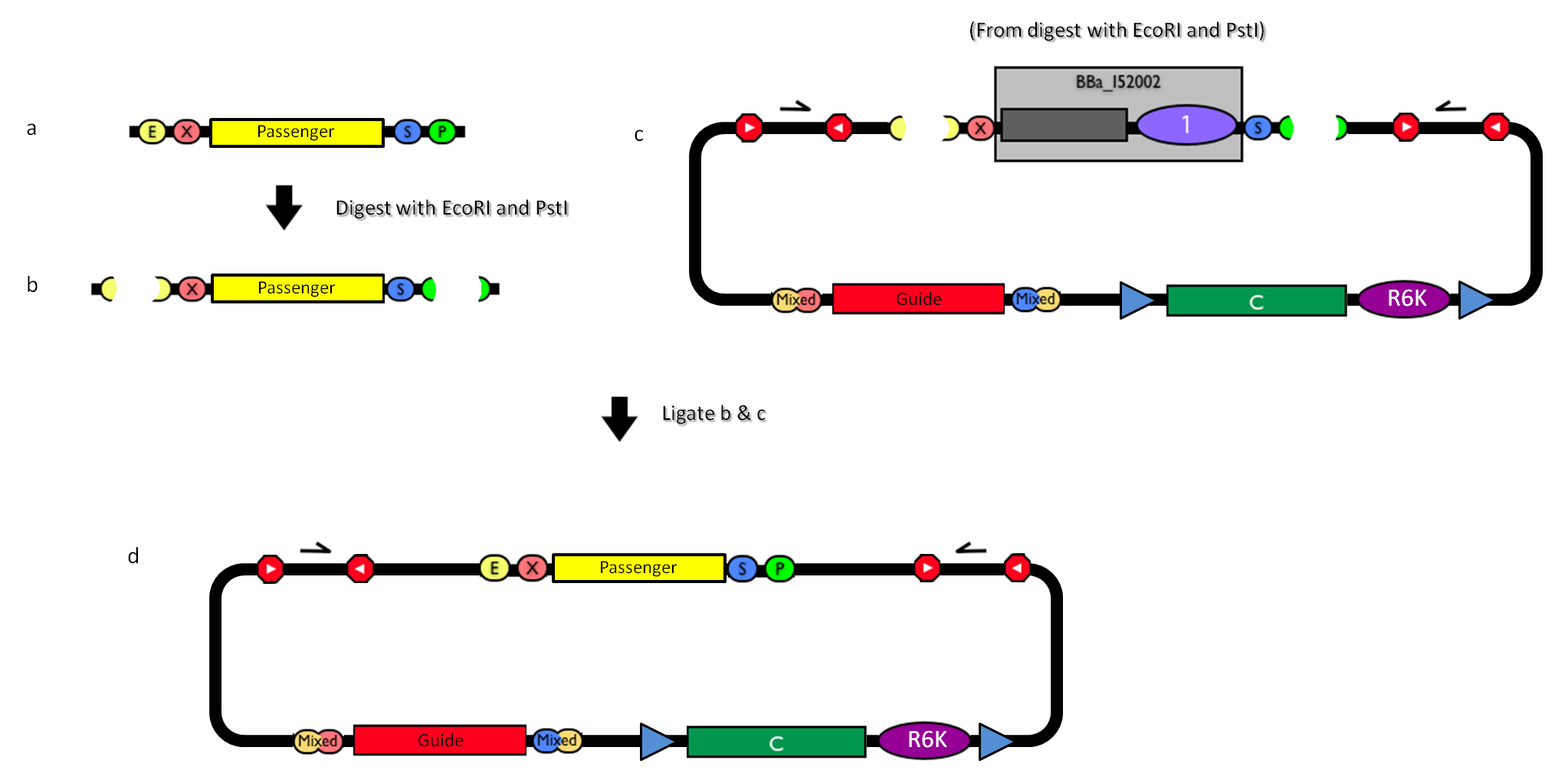
How to engineer itThe DNA guide can be changed as follows:
How to perform genome integrationThe integration into the E. coli chromosome can exploit the bacteriophage attP-mediated integration or the homologous recombination. Detailed protocols about attP-mediated integration can be found here:
Detailed protocols about homologous recombination can be found here:
This integration method is applicable when the host strain does not have prophages in the att(Phi80) locus. TOP10 (<partinfo>BBa_V1009</partinfo>) and DH5alpha (<partinfo>BBa_V1001</partinfo>) strains have the Phi80 prophage and so their chromosome cannot be engineered with this procedure. The genomic integration of the desired BioBrick part into the attP(Phi80) locus has to be mediated by co-transforming a helper plasmid (such as the Amp-resistant <partinfo>BBa_J72008</partinfo> plasmid) carrying the IntPhi80 site-specific integrase gene under the control of a thermoinducible promoter (see Fig.???). The helper plasmid also has a heat-sensitive replication origin, whose replication can be inhibited at temperatures of 37-42°C, while a permissive temperature for this vector is 30°C. For this reason, it can be cured at high temperatures, when the integrase expression is triggered at the same time. The Phi80 integrase mediates the site-specific recombination between the attP site in the integrative vector and the attB site in the bacterial genome (for a schematic description of this process, see Fig.???). Thanks to its R6K conditional replication origin, the integrative vector cannot be replicated in common E. coli strains, so the Chloramphenicol resistant bacteria are actual integrants. In the Materials and Methods section, a detailed protocol to target the desired BioBrick part into the Phi80 locus is reported.
How to perform multiple integration in the same genomeWhen this vector is integrated into the genome, the desired passenger should be maintained into the host, as well as the Chloramphenicol resistance marker and the R6K conditional replication origin. The CmR and the R6K can be excised from the genome by exploiting the two FRT recombination sites that flank them. The Flp recombinase protein mediates this recombination event, so it has to be expressed by a helper plasmid, such as pCP20 (CGSC#7629). This enables the sequential integration of several parts using the same antibiotic resistance marker, which can be eliminated each time. Detailed protocols about homologous recombination can be found here:
Materials and MethodsPlasmids and strains: the <partinfo>BBa_J72008</partinfo> helper plasmid was kindly given by Prof. JC Anderson (UC Berkeley). MC1061 (<partinfo>BBa_K300078</partinfo>) and MG1655 (<partinfo>BBa_V1000</partinfo>) E. coli strains and the pCP20 helper plasmid were purchased from the Coli Genetic Stock Center (Yale University). DH5alpha (<partinfo>BBa_V1001</partinfo>) strain was purchased from Invitrogen. Verification primers: all the oligonucleotides were purchased from Primm (San Raffaele Biomedical Science Park, Milan, Italy). The P1 (<partinfo>BBa_K300975</partinfo>) and P4 (<partinfo>BBa_K300978</partinfo>) primers had already been used in [Anderson JC et al., 2010]. The P2 (<partinfo>BBa_K300976</partinfo>) and P3 (<partinfo>BBa_K300977</partinfo>) primers have been newly designed using ApE and amplifiX. P2 and P3 have been designed also considering the previously used verification primers P2 and P3 in the pG80ko integrative plasmid, described in [DeLoache W, 2009]. Competent cells preparation: all the E. coli strains were made competent following a slightly modified version of the protocol described in [Sambrook J et al., 1989]. Briefly, cells were grown to and OD600 of ~0.4-0.6, harvested (4000 rpm, 10 min, 4°C) and the supernatant discarded. Cells were resuspended in (30 ml for each 50 ml of initial culture) pre-chilled Mg-Ca buffer (80 mM MgCl2, 20 mM CaCl2), centrifuged as before and the supernatant discarded. Cells were resuspended in (2 ml for each 50 ml of initial culture) pre-chilled Ca buffer (100 mM CaCl2, 15% glycerol), aliquoted in 0.5 ml tubes and freezed immediately at -80°C. Test the transformation efficiency as: efficiency [CFU/ug of DNA]= # CFU * 1000 ng of DNA / amount of transformed DNA [ng] Integrative standard vectors for yeastSelf-cleaving affinity tags to easily purify proteins
|
 "
"