Team:ETHZ Basel/Modeling
From 2010.igem.org
(→Mathematical Modeling Overview) |
|||
| Line 6: | Line 6: | ||
]] | ]] | ||
| - | In order to support [[Team:ETHZ_Basel/Biology|wet laboratory experiments]] and to create a test bench for the [[Team:ETHZ_Basel/InformationProcessing|information processing]] part, a complex mathematical model of E. lemming was created. This goal was achieved by implementing and combining deterministic molecular models of the [[Team:ETHZ_Basel/Modeling/Chemotaxis chemotaxis pathway]] and the [[Team:ETHZ_Basel/Modeling/Light_Switch light switch]] and probabilistic model for the [[Team:ETHZ_Basel/Modeling/Movement bacterial movement]]. | + | In order to support [[Team:ETHZ_Basel/Biology|wet laboratory experiments]] and to create a test bench for the [[Team:ETHZ_Basel/InformationProcessing|information processing]] part, a complex mathematical model of E. lemming was created. This goal was achieved by implementing and combining deterministic molecular models of the [[Team:ETHZ_Basel/Modeling/Chemotaxis| chemotaxis pathway]] and the [[Team:ETHZ_Basel/Modeling/Light_Switch| light switch]] and probabilistic model for the [[Team:ETHZ_Basel/Modeling/Movement| bacterial movement]]. |
== Implementation of mathematical models == | == Implementation of mathematical models == | ||
| - | === Individual molecular | + | === Individual molecular models === |
| - | In a first step, we implemented individual molecular models | + | In a first step, we implemented individual deterministic molecular models for the subdevices and a stochastic mathematical model of the bacterial movement. |
| - | + | ||
* [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two similar models of the chemotactic receptor pathway. | * [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two similar models of the chemotactic receptor pathway. | ||
| - | * [[Team:ETHZ_Basel/Modeling/Movement|'''Bacterial Movement''']]: a probabilistic model of E. coli movement, determined by distribution of input bias. | + | * [[Team:ETHZ_Basel/Modeling/Light_Switch|'''Light Switch''']]: based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. |
| + | * [[Team:ETHZ_Basel/Modeling/Movement|'''Bacterial Movement''']]: a probabilistic model of ''E. coli'' movement, determined by distribution of input bias. | ||
=== Combined molecular and mathematical models === | === Combined molecular and mathematical models === | ||
Revision as of 00:25, 22 October 2010
Mathematical Modeling Overview
In order to support wet laboratory experiments and to create a test bench for the information processing part, a complex mathematical model of E. lemming was created. This goal was achieved by implementing and combining deterministic molecular models of the chemotaxis pathway and the light switch and probabilistic model for the bacterial movement.
Implementation of mathematical models
Individual molecular models
In a first step, we implemented individual deterministic molecular models for the subdevices and a stochastic mathematical model of the bacterial movement.
- Chemotaxis Pathway: two similar models of the chemotactic receptor pathway.
- Light Switch: based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3.
- Bacterial Movement: a probabilistic model of E. coli movement, determined by distribution of input bias.
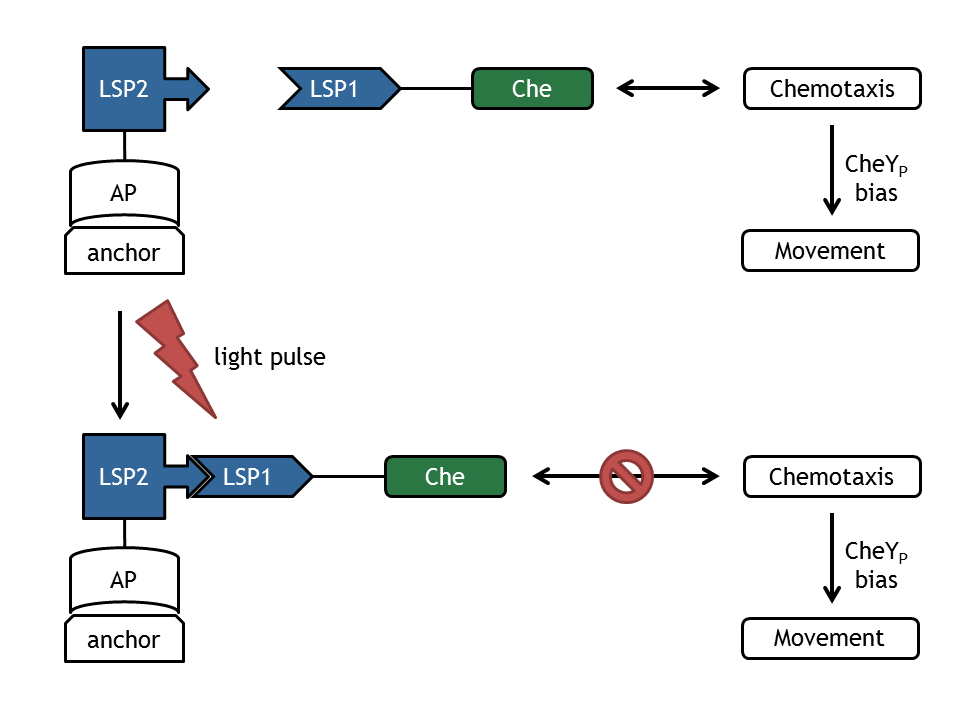
Combined molecular and mathematical models
The next step, combination of the individual molecular and mathematical models to a comprehensive model of E. lemming was achieved in two substeps:
- Light switch - Chemotaxis: used to provide support for wet laboratory.
- Chemotaxis - Movement: complete model of E. lemming.
Experimental Design
Insights for wet laboratory
To create the biological implementation of E. lemming, the parts of the core components had to be chosen in an order to improve chances to result in a functioning ensemble. By using the combined molecular models for in silico evaluation of the best possible parts, it was possible to reduce the amount of different combinations to be tested.
Wet laboratory evaluation results have showed, that molecular modeling and experimental biology can interwork to gain new insight for both perceptions of the problem.
Insights for information processing
In order to adjust the controller to have optimal light pulse rates, the combined molecular model has been used to determine the corresponding time constants.
Information processing evaluation results provide further information how this has been accomplished.
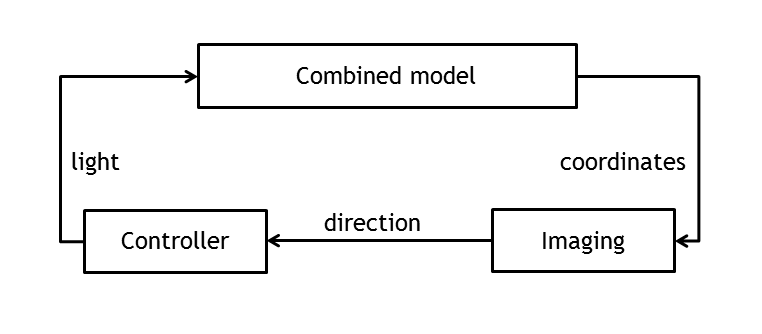
Test bench for information processing
In order to create a first test bench for the information processing pipeline, the combined model has been used to create and evaluate the controller. By providing an input port for light pulse and an output port for bacterial movement, it was possible to close the loop and simulate the whole system.
 "
"