Team:KAIST-Korea/Notebook/Memo/Etc
From 2010.igem.org
| (16 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<table width="100%"> | <table width="100%"> | ||
<tr> | <tr> | ||
| - | <td> | + | <td align=center> |
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Data"><img src="https://static.igem.org/mediawiki/2010/thumb/5/54/Data.jpg/230px-Data.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Data"><img src="https://static.igem.org/mediawiki/2010/thumb/5/54/Data.jpg/230px-Data.jpg"></a></html> | ||
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Info"><img src="https://static.igem.org/mediawiki/2010/thumb/e/e9/Info.jpg/230px-Info.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Info"><img src="https://static.igem.org/mediawiki/2010/thumb/e/e9/Info.jpg/230px-Info.jpg"></a></html> | ||
| Line 15: | Line 15: | ||
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Etc"><img src="https://static.igem.org/mediawiki/2010/thumb/3/39/Etc.jpg/230px-Etc.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Etc"><img src="https://static.igem.org/mediawiki/2010/thumb/3/39/Etc.jpg/230px-Etc.jpg"></a></html> | ||
</td></tr></table> | </td></tr></table> | ||
| - | == <span style=font-size:20px> <b> | + | |
| - | < | + | <table width="100%" border="0" cellpadding="20px"> |
| + | <tr> | ||
| + | <td> | ||
| + | == <span style=font-size:20px> <b> Etc. </b> </span> <span style=font-size:15px>We handle information and idea at ETC </span> == | ||
<br> | <br> | ||
<span style=font-size:15px> <b> MAB </b> </span> | <span style=font-size:15px> <b> MAB </b> </span> | ||
| Line 28: | Line 31: | ||
To use transfomation protocol, if cell growth division gene insert it, it is possible. | To use transfomation protocol, if cell growth division gene insert it, it is possible. | ||
<br><br> | <br><br> | ||
| + | <span style=font-size:15px> <b> Leech anticoagulent </b> </span> | ||
| + | <br> | ||
| + | Long since leech was used to pluck out the blood from people because they though blood reducing relieve the hypertension.<br> | ||
| + | Even now, it is used to relieve swelling of injury.<br> | ||
| + | Leech also have a effect which continues bleeding because it secrets anti-coagulent enzyem. <br> | ||
| + | In mechanical heart valve implementation or thrombosis, to prevent blood clot of vessel people use anti coagulent.<br> | ||
| + | But this obstructs body defense mechanism. If we use leech's anti-coagulent enzyme, we can make medicine. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Hemostasis </b> </span> | ||
| + | <br> | ||
| + | Undesirable clotting is one of disorder of hemostasis.<br> | ||
| + | Thrombus is a clot that develops and persists in an unbroken blood vessel.<br> | ||
| + | Embolus is kind of thrombus that breaks away from the vessel wall and floats freely in the bloodstream.<br> | ||
| + | These can obstruct tiny blood vessel like arteriol or venule.<br> | ||
| + | How about making a bacteria which eliminate them? | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Bacteria Electricity </b> </span> | ||
| + | <br> | ||
| + | Magnetotactic bacteria has a magnet in its body. This bacteria has a magnetosome which makes magnet.<br> | ||
| + | If magnetosome has a regular period and it can make or destroy own magnet, to use change of magnetic field, it can make electricity power.<br> | ||
| + | Actually, this bacteria makes voltage like half of AA battery during 48 hours.<br> | ||
| + | To use this, we can also make electricity. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> ATP Electricity </b> </span> | ||
| + | <br> | ||
| + | Contemporary society's energy source is electricity.<br> | ||
| + | On the other hand, an organism's energy source is ATP.<br> | ||
| + | Such as bio-mass, organism used energy sources researches how chemical energy makes electric energy.<br> | ||
| + | Especially, Making electricity using bio-mass uses method of rolling turbun. But this method are inefficient.<br> | ||
| + | So, from ATP, we can make charge gradient, to use this we can make electric energy. In this case, we don't use living cells.<br> | ||
| + | Just we need ATPase. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Stanford team </b> </span> | ||
| + | <br> | ||
| + | Stanford team's large protein secretion system is hard to use with liposome.<br> | ||
| + | Stanford team combined interleukin6 gene to<br> | ||
| + | :HylA signal sequence<br> | ||
| + | :HylR, HylC, IS1 : Front flanking sequence<br> | ||
| + | :HylB, HylD : Rear flanking sequence<br> | ||
| + | :tolC : channel protein<br> | ||
| + | HylA is secretion inducing sequence, and the others are induced by HylA and make channel to pump proteins to outside of the cell.<br> | ||
| + | It is different from liposome production.<br> | ||
| + | So we need to check another way for liposome based secretion. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Bacterial Secretory Vesicle </b> </span><br> | ||
| + | The appended file says that Bacterial Secretory Vesicle is hard to use. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Cancer diagnosis marker </b> </span><br> | ||
| + | I couldn’t find thesis about cancer diagnosis by bacterial method in NCBI.<br> | ||
| + | It can be both good and bad news.<br> | ||
| + | Attached file is a paper of Prof. Park published in PLoS ONE.<br> | ||
| + | It says that antibody detects surface marker of cancer tissue.<br> | ||
| + | It seems like detecting it by 4 kinds of color reaction.<br> | ||
| + | Maybe we could find another marker from another paper by prof.Lee Do Hyun, for example.<br> | ||
| + | The general concept is to transfer the system to bacteria. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> BBa_K126002 </b> </span><br> | ||
| + | The result of blastx of 'BBa_K126002' is<br> | ||
| + | 'vascular endothelial growth factor receptor 2[Mus musculus](NCBI Reference Sequence: NP_034742.2)'.<br> | ||
| + | And the result of blast of refseq rna is<br> | ||
| + | 'Mus musculus kinase insert domain protein receptor (Kdr), mRNA (NCBI Reference Sequence: NM_010612.2)'.<br> | ||
| + | We now know that the kinase generally come from VEGFR of mouse, so we are searching that the mechanism how VEGFR accerelates growth.<br> | ||
| + | Our purpose is that we intercept the signal of VEGFR's growth accerelating mechanism to induce GFP revelation. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> G-protein Signaling </b> </span><br> | ||
| + | |||
| + | There is a signaling by using G-protein, but Tyrosing kinase.<br> | ||
| + | You could find more information with this address.<br> | ||
| + | :https://2008.igem.org/Team:Illinois/Antibody_GPCR_Fusion<br> | ||
| + | We found the tyrosine receptor's identity using 'BLASTX' and 'refseq-rna DB'. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Y can D pathway </b> </span> | ||
| + | <br> | ||
| + | <table align=center> | ||
| + | <tr><td> | ||
| + | 1. 'Y. canD' is spread on the sample.</td></tr> | ||
| + | <tr><td> | ||
| + | 2. 'Y. canD’s antibody combines with the antigen on the ''Mycobacterium tuberculosis''.</td></tr> | ||
| + | <tr><td> | ||
| + | 3. The tyrosine kinase JAK1, LMO4 and gp130 dimer bind to the cytokine receptor integral to membrane.</td></tr> | ||
| + | <tr><td> | ||
| + | 4. JAK autophosphorylation activates transcription factors called STAT3.</td></tr> | ||
| + | <tr><td> | ||
| + | 5. The activated STAT3 dissociate from the receptor.</td></tr> | ||
| + | <tr><td> | ||
| + | 6. The activated STAT3 form dimers.</td></tr> | ||
| + | <tr><td> | ||
| + | 7. The STAT3 dimer go into nucleus and regulates GFP gene that attached to APRE elements of promoter.</td></tr> | ||
| + | <tr><td> | ||
| + | 8. 'Y.canD' expresse GFP.</td></tr> | ||
| + | </table> | ||
</td></tr></table> | </td></tr></table> | ||
| + | <br><br> | ||
| + | |||
| + | <table width="100%" border="0" cellpadding="20px"> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <span style=font-size:15px> <b> Why Yeast? </b> </span> | ||
| + | <br> | ||
| + | Why we use ‘Yeast’ instead of ‘E. coli’? | ||
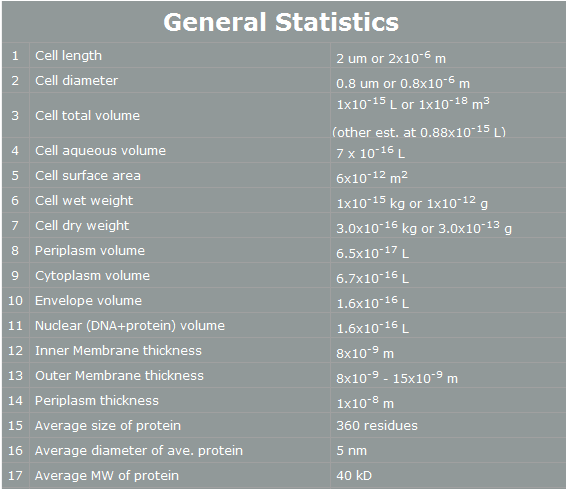
| + | E. coli control is easier than Yeast control. E. coli has faster cell cycle than Yeast. Moreover, there are many E. coli templates. But, although E. coli has many profits, we choose Yeast. Why? The following table is E. coli properties.[2] | ||
| + | <br><br> | ||
| + | [[Image:E.coli general statistics.jpg|500px|center]] | ||
| + | <br> | ||
| + | According to the table, we can find easily some reasons that E. coli cannot work human pathway well.<br><br> | ||
| + | First, the periplasm’s thickness of E. coli is the very small as 10nm. In our project, we have to use human proteins. Almost human proteins are much bigger than E. coli’s proteins. (They are over 10nm*10nm*10nm.) Furthermore, in our project, STAT3 has 60nm height. Therefore, this protein cannot be in periplasm, and our pathway cannot work completely.[3] <br><br> | ||
| + | Second, the average E. coli protein has only 360 residues, 5nm diameter, and 40kD MW. As we mentioned, our project’s proteins have over 1000residues, 10nm diameter and 50kD MW. Thus, the possibility that E. coli have our pathway is very low. <br><br> | ||
| + | However, Yeast has big volume size from 3~4 µm to 40 µm. [4] And Yeast is a eukaryotic, so it is more similar to human cell than E. coli. There is no physical problem of using human protein in yeast. And has more possibility to do our pathway. Therefore, we use Yeast instead of E. coli. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Biochemical Receptors </b> </span> | ||
| + | <br> | ||
| + | [[Image:Int5.png|Biochemical Receptors|center]] | ||
| + | <br> | ||
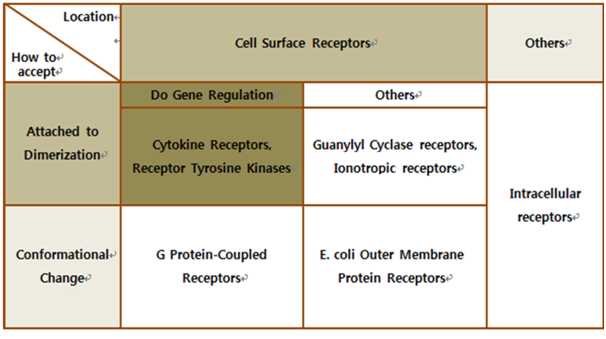
| + | The upper table represents classification of all biological receptors. First of all, since our genetically engineered machine has to detect on mycobacteria directly, we should select cell surface receptors. Next, we should consider using fusion-antibody-receptors. We used gene combination simply (add antibody to biological receptor without original receipt part). If the receptor has conformational change property when it accepts input signal, we guess that our fusion antibody receptor cannot operate well because antibodies characteristics (antigen size, antibody size, etc.) are different. They don’t have special receipt part in a whole receptor, of course, so whole receptor is important receipt part; we cannot create fusion-antibody-receptor with this protein anyway. Therefore, we choose ‘Dimerization’ receptor. Most of these receptors have Immunoglobulin-like receptor part which can be alternated by antibodies easily. Also, since biosensors should activate gene expression, we selected gene-regulation related receptors. <br><br> | ||
| + | Finally we choose cytokine receptors and receptor tyrosine kinases. Among them, interleukin-6 alpha receptor was selected. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Fusion Antibody Receptors </b> </span> | ||
| + | <br> | ||
| + | '''The Fusion Antibody-Receptor''' is a protein which receptor genetically combines with a specific antibody. The following figures show what the fusion antibody-receptor is for details. | ||
| + | <br><br> | ||
| + | [[Image:Int7.png|Fusion Antibody Receptors|center|620px]] | ||
| + | <br> | ||
| + | The fusion antibody-receptor is designed by simple gene modification. We altered the ''Mycobacterium tuberculosis'' antigen detectable antibody single chain instead of the cytokine receptor’s antigen acceptable part (Immunoglobulin-like parts). Light chain part of the antibody – Linker – Heavy chain part of antibody is enough. (There are some evidences to success making fusion antibody-receptor with this method in some experiments. [4]) Then in order to express the receptor on cell surface membrane of Yeast, we change the original signal peptide to Omp(Outer membrane protein) signal peptide of Yeast. Surely the receptor should satisfy our standard for biosensor. | ||
| + | <br> | ||
| + | [[Image:Int8.png|Signal Peptide|center|620px]] | ||
| + | <br> | ||
| + | <b>Schizosaccharomyces pombe Signal peptide</b> from Cell wall integrity and stress response component 1 <br> | ||
| + | |||
| + | Protein sequence: <br> | ||
| + | MVFLNSSPFKGRLLFFVYLLIISTRLVAA <br> | ||
| + | |||
| + | mRNA sequece:<br> | ||
| + | atggtgtttctgaacagcagcccgtttaaaggccgcctgctgttttttgtgtatctgctgattattagcacccgcctggtggcggcg <br> | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Signal Pathway : JAK-STAT Pathway </b> </span> | ||
| + | <br> | ||
| + | There are many signal pathways from plasma membrane receptor to the gene regulation. Figure B is an example of these pathways, MAP kinase pathway linked with RAS pathway. In this pathway, | ||
| + | |||
| + | # With existence of signal molecule, receptor kinases dimerize themselves and phosphorylate each other. | ||
| + | # This phosphorylated receptor dimer is cognized by Grb2 and recruit SOS protein, the Guanine nucleotide Exchange Factor(GEP) | ||
| + | # Recruited SOS protein substitute as GDP of Ras protein with GTP. | ||
| + | # Ras protein with GTP changes its conformation and activate its own kinase activity(Ras pathway so far) | ||
| + | # Ras kinase activate the MAP kinase kinase kinase, MAP KKK(also called as Raf protein) with phosphorylation | ||
| + | # MAP KKK activates the MAP kinase kinase, MAP KK(also called as Mek) with phosphorylation | ||
| + | # MAP KK activates the MAP kinase, MAP K(also called as Erk) with phosphorylation | ||
| + | # Activated MAP Kinase phosphorylates many cytosol proteins. | ||
| + | # For example, if Jun TF is phosphorylated by MAP K, phosphorylated Jun go inside of nucleus and activate the transcription to bind to DNA. (MAP pathway so far) | ||
| + | |||
| + | |||
| + | But to port this signal pathway into the Yeast is not easy because this pathway is related with too many new proteins, while JAK-STAT pathway is easy to port because it doesn’t have many participating proteins. The signal transduction step of JAK-STAT pathway as follows. | ||
| + | <br><br> | ||
| + | [[Image:pathway1.jpg|620px|center]] | ||
| + | <br><br> | ||
| + | # Cytokine binds to the cytokine receptor with JAK kinase. | ||
| + | # Kinases form dimer and are phosphorylated by JAK. | ||
| + | # These phosphorylated tyrosine residue is cognized by STAT protein. | ||
| + | # STAT which cognize phosphorylated tyrosine phosphorylate other STAT protein. | ||
| + | # Two phosphorylated STAT protein form dimer and go inside to the nucleus and activate near gene to bind to the APRE element. | ||
| + | |||
| + | Because JAK-STAT pathway have only 5 steps while RAS-MAP pathway require 9 steps, we decide to port JAK-STAT pathway to the Yeast. | ||
| + | <br><br> | ||
| + | [[Image:pathway2.jpg|620px|center]]<br><br> | ||
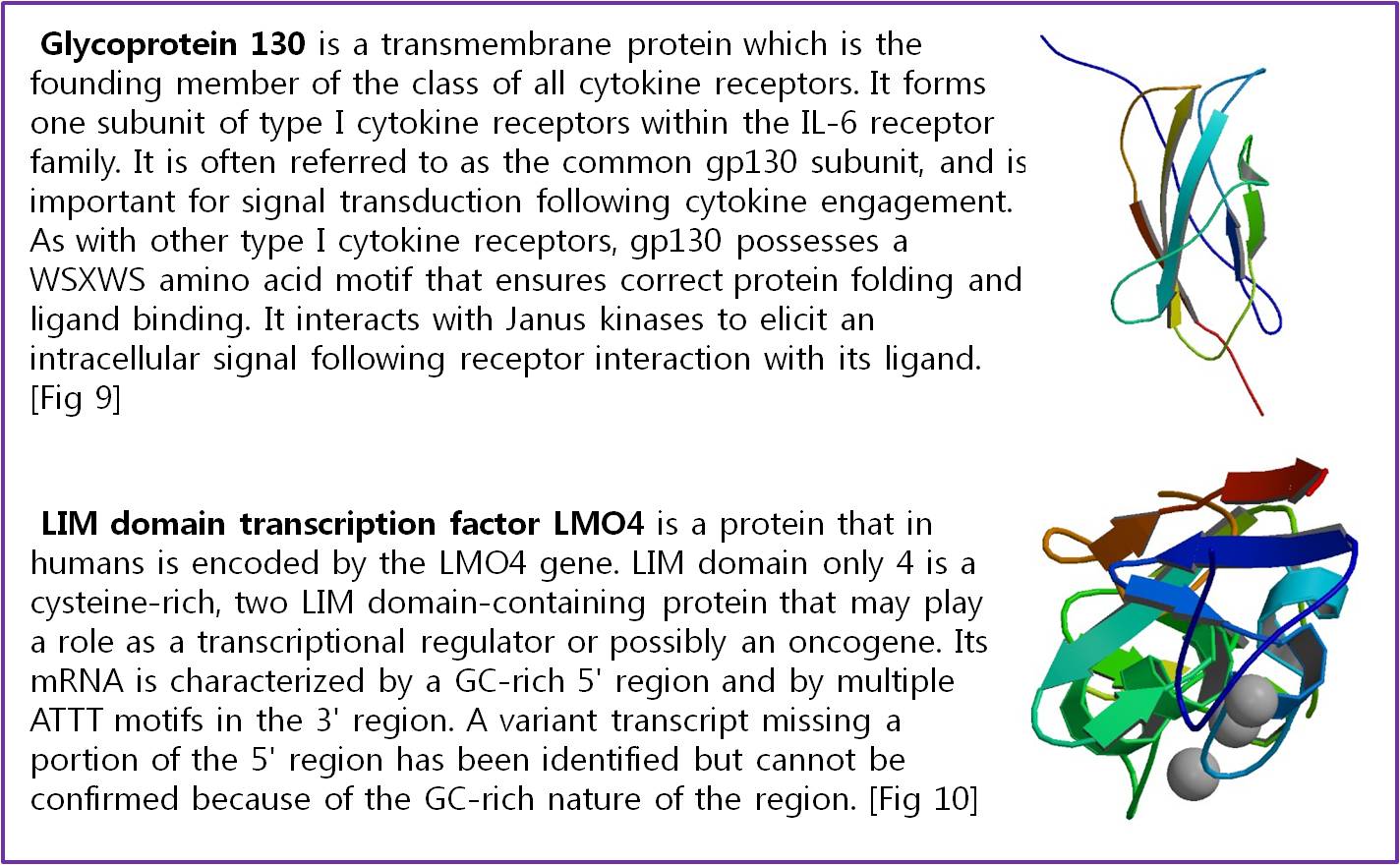
| + | Actually, there are many JAK-STAT pathways in human cell. JAK and STAT are important components of many cytokine receptor systems. According to combination of the receptor, JAK and STAT, there exits many pathways which acts different works in eukaryotic cell. Among these pathways, we choose IL-6α, JAK1, STAT3 pathway. In this pathway, gp130 dimer and LMO4 are combined with JAK1, and assists a whole response. Why we selected the IL-6α specially? That is, IL-6α is a unique receptor which has a Ig-like region at outer part as different with other cytokine receptors. The introduction of IL-6α, gp130, and LMO4 are below. | ||
| + | <br><br> | ||
| + | [[Image:Int6.png|Interleukin Receptors|center|600px]] | ||
| + | <br><br> | ||
| + | [[Image:gp130 LMO4.jpg|620px|center]] | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Gene Activation for Output </b> </span> | ||
| + | <br> | ||
| + | STAT1 and STAT3 proteins with the formation of STAT3/3 and STAT1/3 dimers are confirmed using probe, namely the APRE probe. '''APRE probe'''(''A''cute-''p''hase ''r''esponse ''e''lement probe) is the acute phase responsive element of the α2M gene promoter and known to bind STAT1 and STAT3 proteins. Using this pathway, we altered the '''green fluorescent protein (GFP) gene''' instead of cytokine inducible genes. So, we could show GFP expression through the previous whole pathway.'''[Fig. 5]''' | ||
| + | <br><br> | ||
| + | [[Image:Int9.png|Figure. 5|center]] | ||
| + | <br><br> | ||
| + | |||
| + | <b>FGF signaling pathway</b> | ||
| + | <br> | ||
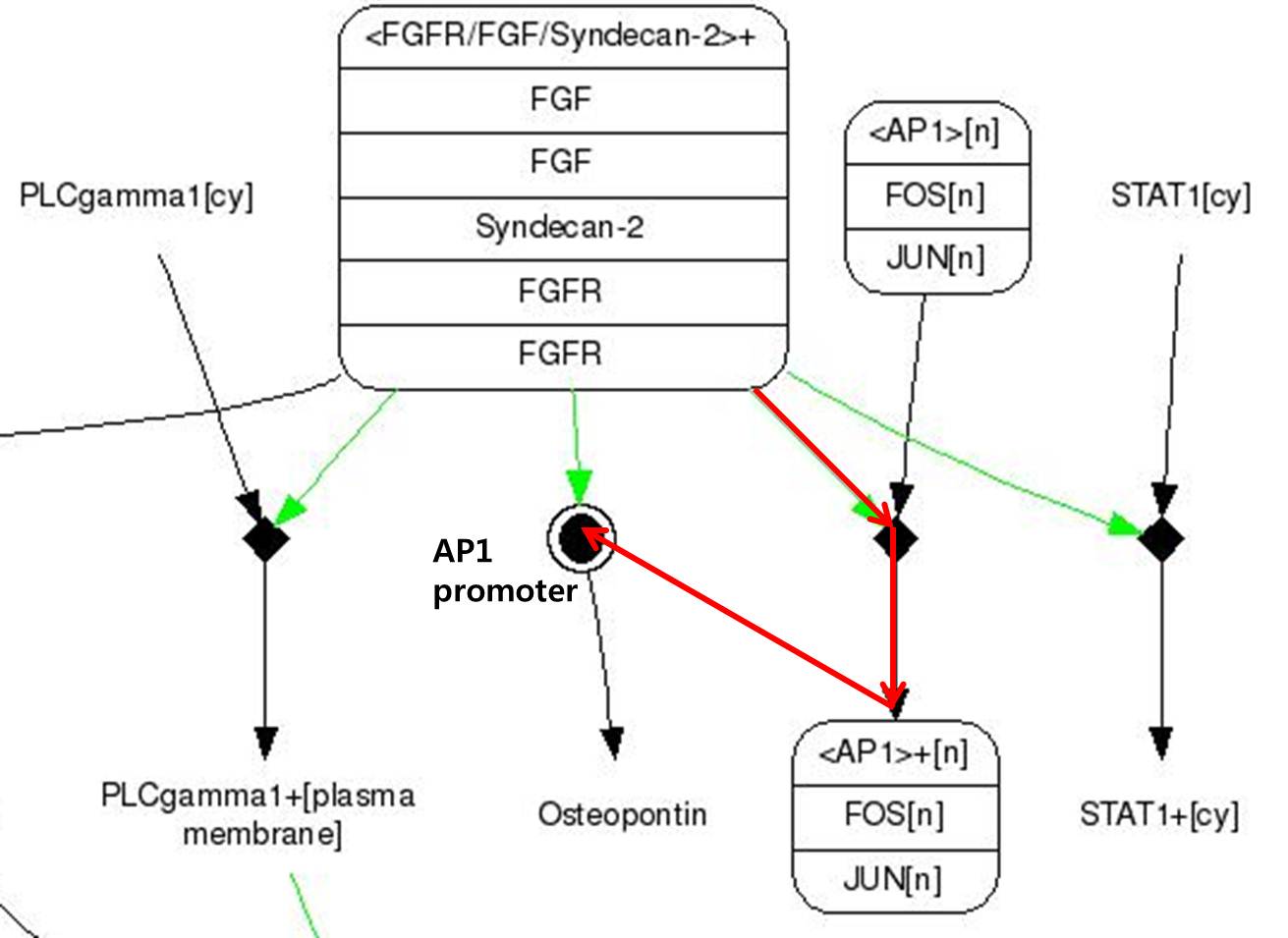
| + | We use FGF signaling pathway(FGF-2) which acts critical role in Craniosynostosis in suture closure. Whole pathway of FGF signaling change osteoblast cell activity eventually. In that one, We focused on osteopontin expression. | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | [[Image:FGF signaling pathway.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 8. FGF signaling Pathway. Red line pathway is selected in our project.</b></center></span> | ||
| + | <br> | ||
| + | |||
| + | # FGF-2s(Fibroblast Growth Factor-2) binds to FGFR(Fibroblast Growth Factor Receptor) | ||
| + | # There are Syndecan-2 which help signal pathway of FGF in membrane. It make FGFR to activate to pass the signal. | ||
| + | # There are FOS and JUN proteins in cytoplasm.(In addtion, FOS and JUN mRNA synthesize more when FGF-2 signal binds.) Especially, c-FOS and c-JUN attach each other and form AP-1(Activate Protein 1). | ||
| + | # Changed FGFR and FGF-2 composition activate AP-1. | ||
| + | # Activated AP-1 can bound to DNA and osteopontin gene is expressd. | ||
| + | <br><br> | ||
| + | |||
| + | Like this,FGF signaling pathways have only 5 steps while RAS-MAP pathway require 9 steps, we decide to port <b>FGF signaling pathway</b> to the Yeast.<br> The introduction of <b>Syndecan-2</b> and <b>AP-1</b> are below. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | [[Image:Syndecan-2.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 9. PDZ-like domain of syndecan-2 </b></center></span> | ||
| + | <br> | ||
| + | [[Image:AP1.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 10. AP-1 (c-FOS + c-JUN) bound to DNA </b></center></span> | ||
| + | <br> | ||
| + | |||
| + | == referance == | ||
| + | :Fig 5. RCSB Protein Database “Crystal structure of the hexameric human IL-6/IL-6 alpha receptor/gp130 complex” | ||
| + | ::http://www.rcsb.org/pdb/explore/explore.do?structureId=1P9M | ||
| + | :Fig 7. Invitrogen “JAK STAT” | ||
| + | ::http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Cell-and-Tissue-Analysis/Signaling-Pathways/Jak-STAT.html | ||
| + | :Fig 8. Akihiko Yoshimura, Hitomi Nishinakamura, Yumiko Matsumura and Toshikatsu Hanada, Negative regulation of cytokine signaling and immune responses by SOCS proteins, Arthritis Research & Therapy(2005) | ||
| + | :Fig 9. RCSB Protein Database “THIRD N-TERMINAL DOMAIN OF GP130” | ||
| + | ::http://www.rcsb.org/pdb/explore/explore.do?structureId=1BJ8 | ||
| + | :Fig 10. RCSB Protein Database “Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain” | ||
| + | ::http://www.rcsb.org/pdb/explore/explore.do?structureId=1RUT | ||
| + | :Fig 11. Roberto, Proteína fluorescente verde – história e perspectivas, Química de Produtos Naturais (2009) | ||
| + | :Fig 9. PDZ-like domain of syndecan-2 | ||
| + | ::RCSB Protein Database, "Solution structure of the atypical PDZ-like domain of synbindin" | ||
| + | :Fig 10. AP-1 (c-FOS + c-JUN) bound to DNA | ||
| + | ::RCSB Protein Database, "STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA " | ||
| + | :[2] Garrett, R.H., and Grisham, C.M. Biochemistry, 2nd Edition (2002), pg. 32</br> | ||
| + | :[3] RCSB PDB(protein database), http://www.rcsb.org/ | ||
| + | :[4] Walker K, Skelton H, Smith K. (2002). accessdate=2009-11-28 "Cutaneous lesions showing giant yeast forms of Blastomyces dermatitidis". Journal of Cutaneous Pathology 29 (10): 616–18. doi:10.1034/j.1600-0560.2002.291009.x | ||
| + | :[5] Dong-Uk Kim et.al, "Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe", Nat.Biotechnol(2010) | ||
| + | :[6] Uniprot Database “Interleukin-6 receptor subunit alpha”; http://www.uniprot.org/uniprot/P08887 | ||
| + | :[7] Lichtman, Andrew H.; Abbas, Abul K. Cellular and molecular immunology (5th ed.). (2003). Philadelphia Saunders. ISBN 0-7216-0008-5. | ||
| + | :[8] SR Roffler1,6, H-E Wang2,6, H-M Yu2, W-D Chang3,4, C-M Cheng3,4, Y-L Lu5, B-M Chen1 and T-L Cheng3,4. A membrane antibody receptor for noninvasive imaging of gene expression. Gene Therapy. Nature (2005) | ||
| + | :[9] Martine I. Darville, Ye-Shih Ho and Decio L. Eizirik, NF-B Is Required for Cytokine-Induced Manganese Superoxide Dismutase Expression in Insulin-Producing Cells, The Endocrine Society(2000) | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
Latest revision as of 05:10, 17 October 2010
|
 "
"