Team:RMIT Australia/Notebook
From 2010.igem.org
SebastianR (Talk | contribs) |
|||
| (55 intermediate revisions not shown) | |||
| Line 405: | Line 405: | ||
</body> | </body> | ||
</html> | </html> | ||
| + | <html> | ||
| + | <style> | ||
| + | table.calendar { margin: 0; padding: 10px; } | ||
| + | table.calendar td { margin: 0; padding: 2px; vertical-align: top; } | ||
| + | table.month .heading td { padding:2px; background-color:#CCCCCC; color:#000000; text-align:center; font-size:120%; font-weight:bold; } | ||
| + | table.month .dow td { color:#3C1859; text-align:center; font-size:110%; } | ||
| + | table.month td.today { background-color:none; } | ||
| + | table.month td { | ||
| + | border: none; | ||
| + | margin: 0; | ||
| + | padding: 1pt 1.5pt; | ||
| + | font-weight: bold; | ||
| + | font-size: 8pt; | ||
| + | text-align: right; | ||
| + | background-color: #eee; | ||
| + | } | ||
| + | #bodyContent table.month a { background:none; padding:0 } | ||
| + | .day-active { color:#FF0000 } | ||
| + | .day-empty { color:#000000 } | ||
| + | </style> | ||
| + | </html> | ||
| + | __NOTOC__ | ||
| + | =='''Notebook'''== | ||
| + | {| align="center" | ||
| + | |-valign="top" | ||
| + | |{{#calendar: title=Team:RMIT_Australia/Notebook# |year=2010 | month=06}} | ||
| + | |{{#calendar: title=Team:RMIT_Australia/Notebook# |year=2010 | month=07}} | ||
| + | |{{#calendar: title=Team:RMIT_Australia/Notebook# |year=2010 | month=08}} | ||
| + | |{{#calendar: title=Team:RMIT_Australia/Notebook# |year=2010 | month=09}} | ||
| + | |{{#calendar: title=Team:RMIT_Australia/Notebook# |year=2010 | month=10}} | ||
| + | |} | ||
| + | {| | ||
| + | |-valign="top" border="0" | ||
| + | |width="100%" style="padding: 0 20px 0 0;"| | ||
| - | |||
| - | |||
| - | + | {|style="text-align:center"; width="100%"; "font-size:large" | |
| - | + | |- | |
| + | |-valign="top" | ||
| + | |width="50%" | | ||
| + | <div id=" .2F1_May_2010"></div> | ||
| + | <div id=" .2F2_May_2010"></div> | ||
| + | <div id=" .2F3_May_2010"></div> | ||
| + | <div id=" .2F4_May_2010"></div> | ||
| + | <div id=" .2F5_May_2010"></div> | ||
| + | <div id=" .2F6_May_2010"></div> | ||
| + | <div id=" .2F7_May_2010"></div> | ||
| + | <div id=" .2F8_May_2010"></div> | ||
| + | <div id=" .2F9_May_2010"></div> | ||
| + | <div id=" .2F10_May_2010"></div> | ||
| + | <div id=" .2F11_May_2010"></div> | ||
| + | <div id=" .2F12_May_2010"></div> | ||
| + | <div id=" .2F13_May_2010"></div> | ||
| + | <div id=" .2F14_May_2010"></div> | ||
| + | <div id=" .2F15_May_2010"></div> | ||
| + | <div id=" .2F16_May_2010"></div> | ||
| + | <div id=" .2F17_May_2010"></div> | ||
| + | <div id=" .2F18_May_2010"></div> | ||
| + | <div id=" .2F19_May_2010"></div> | ||
| + | <div id=" .2F20_May_2010"></div> | ||
| + | <div id=" .2F21_May_2010"></div> | ||
| - | === | + | <div id=" .2F28_June_2010"> |
| + | ===28 June=== | ||
| - | + | Did OH&S training and PC2 training | |
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | <div id=" .2F29_June_2010"> | |
| - | + | ===29 June=== | |
| - | + | ||
| - | + | Began research on what we were going to use to promote taq | |
| + | </div> | ||
| - | + | <div id=" .2F30_June_2010"> | |
| - | + | ===30 June=== | |
| - | + | ||
| - | + | Looked at T7 with a Lac operon as a promotore of taq | |
| + | </div> | ||
| - | + | <div id=" .2F13_July_2010"> | |
| + | ===13 July=== | ||
| - | + | Designed Primers for Taq into backbone | |
| - | + | Researched about using an Asp/Pro between the peptide and the Taq | |
| - | + | Began looking at ligation independent cloning for insertion of peptide | |
| - | + | </div> | |
| - | + | <div id=" .2F14_July_2010"> | |
| + | ===14 July=== | ||
| - | + | Looked at prices of Hotels in Boston | |
| - | + | </div> | |
| - | + | <div id=" .2F19_July_2010"> | |
| - | + | ===19 July=== | |
| + | began looking at the structure of taq, its thermostability | ||
| + | began looking at different mutations that can be done to taq to increase its thermostability without changing its structure | ||
| - | + | </div> | |
| + | <div id=" .2F21_July_2010"> | ||
| + | ===21 July=== | ||
| + | sent in sequence of wild type taq to iTasser | ||
| - | + | </div> | |
| - | + | <div id=" .2F23_July_2010"> | |
| - | + | ===23 July=== | |
| - | + | ||
| + | Checked thermostability of the mutations that will be done to Taq using Expasy | ||
| - | + | Sent in sequence of taq with mutations at 485, 515 and 540 to iTasser | |
| + | |||
| + | <div id=" .2F3_August_2010"> | ||
| + | ===3 August=== | ||
| - | + | Had iGEM team meeting | |
| - | + | -Discussed ANZAAS Proposal | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -Spoke about possibilities of using Geneart | |
| - | + | -Discussed possible T-shirt designs | |
| - | === | + | </div> |
| + | <div id=" .2F5_August_2010"> | ||
| + | ===5 August=== | ||
| - | + | Meet up with Damian, discussed different strategies to use in modelling, began using YASARA to do our Modelling | |
| - | + | </div> | |
| - | + | <div id=" .2F10_August_2010"> | |
| + | ===10 August=== | ||
| - | + | Had iGEM team meeting | |
| - | + | -Took Team Photo | |
| - | + | </div> | |
| + | <div id=" .2F11_August_2010"> | ||
| + | ===11 August=== | ||
| - | The | + | PCR of Taq |
| + | |||
| + | Taq with a DNA concentration of 40ng/µL was diluted to 1µM by addition of 24µL of MilliQ water to 1µL of the Taq plasmid. Primers were diluted to 100µM by adding 354µL to the forward primer with a concentration of 35.4 nmol and 405µL to the reverser primer with a concentration of 40.5nmol. The primers were further diluted to 0.2µM by adding 10µL of the primer to 490µL of milliQ water. The PCR reaction was then made by adding 25µL of Master Mix, 2.5µL of each primer, 5µL of Rnase free water and 15µL of 0.2µM Taq Plasmid. | ||
| + | |||
| + | The PCR | ||
| + | |||
| + | 98º 60 Sec 1 cycle | ||
| + | |||
| + | 98º 15 Sec then 72º 90 Sec 25 cycle | ||
| + | |||
| + | 72º 5 min 1 cycle | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F16_August_2010"> | ||
| + | |||
| + | ===16 August=== | ||
| + | |||
| + | Collaborated with the Sheffield team by participating in an over the phone survery about Human Practices | ||
| + | |||
| + | </div> | ||
| + | <div id=" .2F17_August_2010"> | ||
| + | ===17 August=== | ||
| + | |||
| + | |||
| + | had team meeting | ||
| + | |||
| + | - discussed how we can get more sponsors | ||
| + | |||
| + | - talked about placing our order to gene art | ||
| + | |||
| + | - began filling out the form to send Pennstates survey to the ethics committee | ||
| + | |||
| + | - discussed results for modelling | ||
| + | |||
| + | - decided to fix optimise our previous PCR | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F18_August_2010"> | ||
| + | ===18 August=== | ||
| + | |||
| + | called few companies seeking sponsorship | ||
| + | enquired for booking flights and accomadation | ||
| + | |||
| + | </div id=" .2F19_August_2010"> | ||
| + | ===19 August=== | ||
| + | |||
| + | Working on comparing TAQ wild structure to the TAQ Mutated structure to see that there are no major differences, which will mean that TAQ is inactive but will retain it's thermostability at high temperatures. | ||
| + | |||
| + | Wrote an email to ABC TV, wanting to know if they will do piece on our project for there The New Inventors Program. | ||
| + | |||
| + | Waiting for results from I-Tasser on the mutated TAQ sequence. | ||
| + | Went into the lab and made SOC media and LB- agar plates | ||
| + | |} | ||
| + | |||
| + | <div id=" .2F20_August_2010"> | ||
| + | ===20 August=== | ||
| + | |||
| + | Did two PCR reaction and a transformation | ||
| + | The first PCR we ran was a touch down PCR. Temperatures ranged from 69 to 64. | ||
| + | The second PCR was a two step PCR. | ||
| + | |||
| + | <div id=" .2F30_August_2010"> | ||
| + | ===30 August=== | ||
| + | |||
| + | Ordered the TAQ polymerase from Mr. Gene. Should have it in 15 days. | ||
| + | Meeting today at 10am to discuss project budget and costings. | ||
| + | After the meeting we all are sitting together for a writing session. | ||
| + | |||
| + | <div id=" .2F31_August_2010"> | ||
| + | ===31 August=== | ||
| + | |||
| + | Spent the morning organising our flights | ||
| + | In the afternoon we all sat together for a writing session | ||
| + | |||
| + | <div id=" .2F1_September_2010"> | ||
| + | ===1 September=== | ||
| + | |||
| + | We all sat together for another writing session. | ||
| + | |||
| + | <div id=" .2F7_September_2010"> | ||
| + | ===7 September=== | ||
| + | |||
| + | Had a team meeting | ||
| + | |||
| + | <div id=" .2F14_September_2010"> | ||
| + | ===14 September=== | ||
| + | |||
| + | Had a team meeting and made a lab time table, we also set out tasks which need to be done by the next meeting | ||
| + | *Update the wiki | ||
| + | |||
| + | * Finalise the form for the ethics committee for pennstate | ||
| + | |||
| + | * Call up possible sponsors | ||
| + | |||
| + | * Collect Tshirts | ||
| + | |||
| + | * Work on abstract and team Roster | ||
| + | |||
| + | * research heat liable bonds | ||
| + | |||
| + | * order peptides | ||
| + | |||
| + | * chat with emilio about possible bio assays | ||
| + | |||
| + | * work on making the promotor part | ||
| + | |||
| + | <div id=" .2F15_September_2010"> | ||
| + | ===15 September=== | ||
| + | |||
| + | '''Mutagenesis Quikchange XL kit''' | ||
| + | |||
| + | [https://2010.igem.org/Team:RMIT_Australia/Notebook/Mutagenesis Click Here for Mutagenesis Protocol] | ||
| + | |||
| + | *Template Concentration = 8ng/µl | ||
| + | |||
| + | *Fwd 77ng/µl (low 260/280) | ||
| + | *Rev 86.5 ng/µl (low 260/280) | ||
| + | We optimised for low 260/280 | ||
| + | |||
| + | '''Reaction Mixture''' | ||
| + | *5 µl buffer | ||
| + | *1.9 µl template | ||
| + | *1.7 µl Fwd | ||
| + | *1.62 µl Rev | ||
| + | *1 µl dNTP | ||
| + | *38.78 µl H2O --> reaction volume 50 µl | ||
| + | *1 µl Pfu | ||
| + | |||
| + | First mutagenesis was to turn the -35 site in the promoter region into an Afi II restriction site. | ||
| + | Dpn1 digest (2 µl) for 2hrs | ||
| + | |||
| + | <div id=" .2F16_September_2010"> | ||
| + | ===16 September=== | ||
| + | |||
| + | [https://2010.igem.org/Team:RMIT_Australia/Notebook/Mutagenesis Click Here for Mutagenesis Protocol] | ||
| + | |||
| + | 2 µl of the mutagenesis product into 100 µl of competent cells | ||
| + | Plate 50 µl, 100 µl and 150 µl samples. | ||
| + | |||
| + | <div id=" .2F17_September_2010"> | ||
| + | |||
| + | ===17 September=== | ||
| + | |||
| + | - finalized project abstract | ||
| + | |||
| + | <div id=" .2F19_September_2010"> | ||
| + | ===19 September=== | ||
| + | |||
| + | |||
| + | - Sent off project abstract to HQ | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F20_September_2010"> | ||
| + | ===20 September=== | ||
| + | |||
| + | Two colonies were picked from the plates and were grown in 5mL of Media | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id=" .2F21_September_2010"> | ||
| + | ===21 September=== | ||
| + | |||
| + | A miniprep was performed on 1.5ul of the overnight culture. | ||
| + | 10ng/ul DNA were obtained from both samples with a O.D higher than 1.85. These samples were then used to perform a restriction digest using EcoR1 H.F and AflII. The restriction digest was done according to the iGEM protocols [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.pdf CLICK HERE]. | ||
| + | |||
| + | '''Results''' | ||
| + | The restriction digest did not work. We expected a band at 160bp and one at ~3000bp | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F22_September_2010"> | ||
| + | ===22 September=== | ||
| + | |||
| + | >>>> TABLE <<<< | ||
| + | |||
| + | |||
| + | '''Results''' | ||
| + | The restriction worked. Gel Image below | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F23_September_2010"> | ||
| + | ===23 September=== | ||
| + | Second mutagenesis. This involves changing the -10 promoter region into a ClaI restriction site. AflII (11) miniprep sample was used as template | ||
| + | |||
| + | [https://2010.igem.org/Team:RMIT_Australia/Notebook/Mutagenesis Click Here for Mutagenesis Protocol] | ||
| + | |||
| + | After the Dpn1 digest, 2ul of product was transformed into E.coli Xl1 blue cells. | ||
| + | </div> | ||
| + | <div id=" .2F24_September_2010"> | ||
| + | ===24 September=== | ||
| + | Colonies grew on the plate. This was stores in the fridge (4 degrees) until Monday for colonies to be picked. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F27_September_2010"> | ||
| + | ===27 September=== | ||
| + | Two colonies were picked from the plate and grown overnight at 37 degrees in the shaking incubator. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F28_September_2010"> | ||
| + | ===28 September=== | ||
| + | Preparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) | ||
| + | |||
| + | Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F29_September_2010"> | ||
| + | ===29 September=== | ||
| + | Two more colonies (different) were picked and grown overnight at 37 degrees. | ||
| + | |||
| + | Transformation of RFP in Ampicillin, Kanamycin and Chloroamphenicol backbones. They will act as spare backbones. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F30_September_2010"> | ||
| + | |||
| + | ===30 September=== | ||
| + | A second preparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) | ||
| + | |||
| + | Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture again. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. There is a problem with the backbone or the miniprep. Must investigate. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F4_October_2010"> | ||
| + | ===4 October=== | ||
| + | |||
| + | Got it! | ||
| + | The problem was the plasmid backbone. It was thought that it was pSB1A3, but in fact it was pSB1AK3. Kanamycin has a ClaI site. | ||
| + | |||
| + | A restriction digest was performed using EcoRI and PstI. The RFP plasmid was also digested with EcoRI and PstI. A ligation reaction was performed in order for the promoter part to switch backbones (from pSB1AK3 to pSB1C3). | ||
| + | RFP is also a form of control mechanism to see if the plasmid has the correct insert. Red colonies have RFP, while White colonies have our insert. Overnight ligation at 16 degrees. | ||
| + | |||
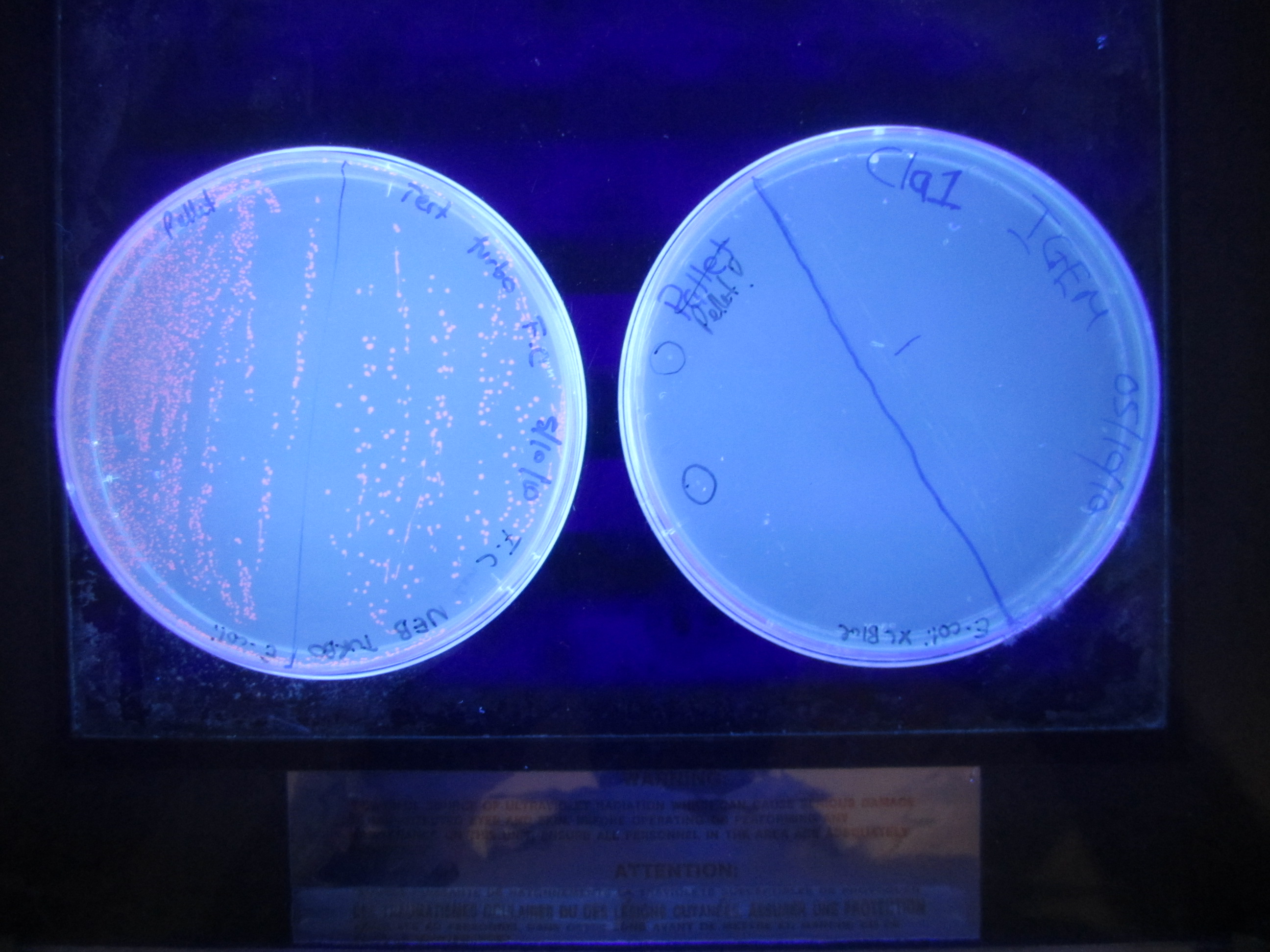
| + | [[Image:RedWhite.JPG| 350px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id=" .2F5_October_2010"> | ||
| + | |||
| + | ===5 October=== | ||
| + | |||
| + | The ligation product was transformed (5ul) into E.coli XL1 Blue competent cells. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id=" .2F6_October_2010"> | ||
| + | ===6 October=== | ||
| + | |||
| + | Colonies grew. Two white colonies were picked and grown overnight in the shaking incubator at 37 degrees. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F7_October_2010"> | ||
| + | ===7 October=== | ||
| + | |||
| + | Slow growth on Chloroamphenicol. Cultures were left on the shaking incubator for an extra night. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F8_October_2010"> | ||
| + | ===8 October=== | ||
| + | |||
| + | Miniprep of the colonies. One successfully grew, while the other did not. | ||
| + | Miniprep results gave a concentration of 114ng/ul with a 260/280 of 1.98. Woohoo. | ||
| + | |||
| + | Double digest (diagnostic) with NcoI and AflII/ClaI --> to check the success of mutagenesis. Gel result gave strange band pattern. | ||
| + | </div> | ||
| + | |||
| + | <div id=" .2F11_October_2010"> | ||
| + | ===11 October=== | ||
| + | |||
| + | Single restriction digest (AflII/ClaI) verified the size of the plasmid. Will re-do double digest tomorrow. | ||
| + | |||
| + | working on presentation and wiki | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id=" .2F13_October_2010"> | ||
| + | ===13 October=== | ||
| + | Making 80% glycerol stock | ||
| + | Miniprep of mr.gene TAQ | ||
| + | clone 1 = 26.7 ng/ul (100ul) | ||
| + | clone 2 = 21.1ng/ul (100ul) | ||
| + | Restriction Enzyme digest of mr.gene taq with EcoRi and Pst | ||
| + | Diagnostic Gel worked YAY | ||
| + | Ligation of Mr.gene taq into psb1c3 left over night at 16 degrees | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div id=" .2F14_October_2010"> | ||
| + | ===14 October=== | ||
| + | Heat inactivated T4 ligase | ||
| + | Transformation of Mr.gene taq in psb1c3 | ||
| + | </div> | ||
Latest revision as of 05:06, 14 October 2010


Notebook
|
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
20 AugustDid two PCR reaction and a transformation The first PCR we ran was a touch down PCR. Temperatures ranged from 69 to 64. The second PCR was a two step PCR. 30 AugustOrdered the TAQ polymerase from Mr. Gene. Should have it in 15 days. Meeting today at 10am to discuss project budget and costings. After the meeting we all are sitting together for a writing session. 31 AugustSpent the morning organising our flights In the afternoon we all sat together for a writing session 1 SeptemberWe all sat together for another writing session. 7 SeptemberHad a team meeting 14 SeptemberHad a team meeting and made a lab time table, we also set out tasks which need to be done by the next meeting
15 SeptemberMutagenesis Quikchange XL kit Click Here for Mutagenesis Protocol
We optimised for low 260/280 Reaction Mixture
First mutagenesis was to turn the -35 site in the promoter region into an Afi II restriction site. Dpn1 digest (2 µl) for 2hrs 16 SeptemberClick Here for Mutagenesis Protocol 2 µl of the mutagenesis product into 100 µl of competent cells Plate 50 µl, 100 µl and 150 µl samples. 17 September- finalized project abstract 19 September- Sent off project abstract to HQ 20 SeptemberTwo colonies were picked from the plates and were grown in 5mL of Media 21 SeptemberA miniprep was performed on 1.5ul of the overnight culture. 10ng/ul DNA were obtained from both samples with a O.D higher than 1.85. These samples were then used to perform a restriction digest using EcoR1 H.F and AflII. The restriction digest was done according to the iGEM protocols [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.pdf CLICK HERE]. Results The restriction digest did not work. We expected a band at 160bp and one at ~3000bp 22 September>>>> TABLE <<<<
23 SeptemberSecond mutagenesis. This involves changing the -10 promoter region into a ClaI restriction site. AflII (11) miniprep sample was used as template Click Here for Mutagenesis Protocol After the Dpn1 digest, 2ul of product was transformed into E.coli Xl1 blue cells. 24 SeptemberColonies grew on the plate. This was stores in the fridge (4 degrees) until Monday for colonies to be picked. 27 SeptemberTwo colonies were picked from the plate and grown overnight at 37 degrees in the shaking incubator. 28 SeptemberPreparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. 29 SeptemberTwo more colonies (different) were picked and grown overnight at 37 degrees. Transformation of RFP in Ampicillin, Kanamycin and Chloroamphenicol backbones. They will act as spare backbones. 30 SeptemberA second preparation of plasmid DNA using Invitrogen Miniprep Kit. Obtained ---ng/ul (260/280 = ---) Restriction digest using EcoRI and ClaI, then gel electrophoresis. Strange gel picture again. Two bands of similar size ~ 1.3 kb and 1.6 kb appear on the gel. There is a problem with the backbone or the miniprep. Must investigate. 4 OctoberGot it! The problem was the plasmid backbone. It was thought that it was pSB1A3, but in fact it was pSB1AK3. Kanamycin has a ClaI site. A restriction digest was performed using EcoRI and PstI. The RFP plasmid was also digested with EcoRI and PstI. A ligation reaction was performed in order for the promoter part to switch backbones (from pSB1AK3 to pSB1C3). RFP is also a form of control mechanism to see if the plasmid has the correct insert. Red colonies have RFP, while White colonies have our insert. Overnight ligation at 16 degrees. 5 OctoberThe ligation product was transformed (5ul) into E.coli XL1 Blue competent cells. 6 OctoberColonies grew. Two white colonies were picked and grown overnight in the shaking incubator at 37 degrees. 7 OctoberSlow growth on Chloroamphenicol. Cultures were left on the shaking incubator for an extra night. 8 OctoberMiniprep of the colonies. One successfully grew, while the other did not. Miniprep results gave a concentration of 114ng/ul with a 260/280 of 1.98. Woohoo. Double digest (diagnostic) with NcoI and AflII/ClaI --> to check the success of mutagenesis. Gel result gave strange band pattern. 11 OctoberSingle restriction digest (AflII/ClaI) verified the size of the plasmid. Will re-do double digest tomorrow. working on presentation and wiki 13 OctoberMaking 80% glycerol stock Miniprep of mr.gene TAQ clone 1 = 26.7 ng/ul (100ul) clone 2 = 21.1ng/ul (100ul) Restriction Enzyme digest of mr.gene taq with EcoRi and Pst Diagnostic Gel worked YAY Ligation of Mr.gene taq into psb1c3 left over night at 16 degrees
14 OctoberHeat inactivated T4 ligase Transformation of Mr.gene taq in psb1c3 |
 "
"