Team:Cambridge/References/ProjectBioluminescence/Luciferase
From 2010.igem.org
(Redirected from Team:Cambridge/ProjectBioluminescence/Luciferase)

Link dumps: Bioluminescence |
Firefly luciferases|
Lucferin recovery|
Light output|
Experiments|
Modelling
Bioluminescence: Luciferases
Mutant luciferase (P. pyralis)
- [http://www.ncbi.nlm.nih.gov/nuccore/AB261988.1 Mutant Enhanced Photon Initiating Complex (EPIC) Luciferase (P.pyralis) Sequence]
- [http://www.ncbi.nlm.nih.gov/pubmed/10529195 Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color]
- [http://www.ncbi.nlm.nih.gov/pubmed/12044905 Improved practical usefulness of firefly luciferase by gene chimerization and random mutagenesis]
Other Insect luciferases
- For colours we could use the click beetle luciferase [http://partsregistry.org/wiki/index.php/Part:BBa_J70005 red] or [http://partsregistry.org/wiki/index.php/Part:BBa_J70006 green]. They're only at the 'planning' stage though.
Bacterial Bioluminescence
- [http://arjournals.annualreviews.org/doi/pdf/10.1146/annurev.mi.42.100188.001055?cookieSet=1 paper with hard numbers, pathways]
- [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC372803/pdf/microrev00032-0137.pdf The definitive summary paper]
- Vibrio Fischeri
- Vibrio Harveyi
- Vibrio Phosphoreum
- Photorhabdus
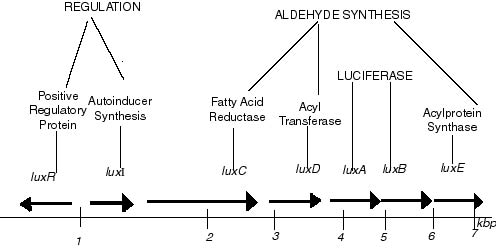
- Genetics of luxCDABE (adapted from wikipedia)
- 2006 attempts at BioBricking the lux operon
- Notes
- [http://partsregistry.org/Lux The lux wiki from parts registry]
- [http://www.ncbi.nlm.nih.gov/nuccore/AF170104.1?report=graph&log$=seqview NCBI Lux operon sequence]
- [http://partsregistry.org/Part:BBa_G10001 parts registry lux operon] -currently waiting on an email to see how useful/available this part is
- IMPORTANT INFO
- The Photorhabdus luminescens [http://partsregistry.org/Part:BBa_J70008 luxCDABE operon] in the registry. It's not well characterised
Plasmid experiment
- [http://departments.kings.edu/biology/lux/bacterial.html Vibrio plasmid experiment] -possible source of lux operon
- [http://departments.kings.edu/biology/lux/index.html] - Transformation protocol for students from basics - includes several plasmids
- Plan
- set plasmid 555 into a BioBrick
- replace the promoter? sensitivity tuners?
- references
- [http://www.pnas.org/content/84/19/6639.short Overproduction and purification of the luxR gene product: Transcriptional activator of the Vibrio fischeri luminescence system (Kaplan & Greenburg 1987) ]
- [http://jb.asm.org/cgi/content/abstract/172/7/3974 Critical regions of the Vibrio fischeri luxR protein defined by mutational analysis (Slock et al 1990)]
Transformation Experiment to test Firefly luciferase in registry
- Firefly luciferase is in the registry as [http://partsregistry.org/Part:BBa_I712019 BBa_I712019], and have been sent to us as part of the spring 2010 distribution.
- We need to find a new vector with a bacterial ribosyme binding site, initiator, and a stop codon. Having searched via the catalog in the plasmids section, then in expression plasmids, we have selected a constitutive plasmid:
- The plasmid selected is [http://www.partsregistry.org/Part:BBa_J13002 BBa_J13002], which is a TetR repressed POPS/RIPS generator, again part of the spring 2010 distribution - kit plate 1, well 13B, plasmid pSB1A2. It was proven to work by UMN iGEM 2009.
Wavelengths
548 nm http://pubs.acs.org/doi/pdf/10.1021/bi7015052
613 nm http://www.promega.com/pnotes/85/10904_11/10904_11.pdf
| Luciferase | Colour(at pH7.8) | λmax(nm) | (pH 7.8) | (pH 6.0) | Base change | Amino acid change | |
|---|---|---|---|---|---|---|---|
| Genjii | yellow-green | 562 | 609 | ||||
| C-M-1 | orange | 607 | 614 | G857-->A | Ser286-->Asn | ||
| C-M-2 | red | 609 | 611 | G976-->A | Gly326-->Ser | ||
| C-M-3 | red | 612 | 612 | C1297-->T | His433-->Tyr | ||
| C-M-4 | yellow-orange | 595 | 609 | C1354-->T | Pro452-->Ser | ||
| C-M-6 | green | 558 | 558 | G715-->A | Val239-->Ile | ||
| C-M-11 | yellow | 565 | 612 |
 "
"