Template:UNIPV-Pavia/Project/results/Self-cleaving affinity tags to easily purify proteins
From 2010.igem.org
(→Methods) |
(→Results) |
||
| Line 21: | Line 21: | ||
===Results=== | ===Results=== | ||
====5 hours'==== | ====5 hours'==== | ||
| - | [[Team:UNIPV-Pavia/Material_Methods/Protocols#Sudan_Black_staining_protocol|Sudan Black staining protocol]] was performed on 70ul | + | [[Team:UNIPV-Pavia/Material_Methods/Protocols#Sudan_Black_staining_protocol|Sudan Black staining protocol]] was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here: |
{|align="center" | {|align="center" | ||
| Line 27: | Line 27: | ||
|[[Image:UNIPV10_RBS32_nothing_5h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 5 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2_5h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 5 hours (negative control)]] | |[[Image:UNIPV10_RBS32_nothing_5h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 5 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2_5h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 5 hours (negative control)]] | ||
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_nothing5h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_nothing5h.jpg|thumb|200px|center|DSMZ15372 with nothing added in the culture, after 5 hours]] || |[[Image:UNIPV10_PBHR68_nothing_2_5h.jpg|thumb|200px|center|DSMZ15372 with nothing added in the culture, after 5 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_gly_5h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_gly_5h.jpg|thumb|200px|center|DSMZ15372 with 2% glycerol added in the culture, after 5 hours]] || |[[Image:UNIPV10_PBHR68_gly_2_5h.jpg|thumb|200px|center|DSMZ15372 with 2% glycerol added in the culture, after 5 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_IPTG_5h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_IPTG_5h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG added in the culture, after 5 hours]] || |[[Image:UNIPV10_PBHR68_IPTG_2_5h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG in the culture, after 5 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_gly_IPTG_5h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_gly_IPTG_5h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG and 2% glycerol added in the culture, after 5 hours]] || |[[Image:UNIPV10_PBHR68_gly_IPTG_2_5h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG and 2% glycerol added in the culture, after 5 hours]] |
|} | |} | ||
| Line 39: | Line 39: | ||
====30 hours'==== | ====30 hours'==== | ||
| - | [[Team:UNIPV-Pavia/Material_Methods/Protocols#Sudan_Black_staining_protocol|Sudan Black staining protocol]] was performed on 70ul | + | [[Team:UNIPV-Pavia/Material_Methods/Protocols#Sudan_Black_staining_protocol|Sudan Black staining protocol]] was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here: |
{|align="center" | {|align="center" | ||
| Line 45: | Line 45: | ||
|[[Image:UNIPV10_RBS32_nothing_30h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2_30h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] | |[[Image:UNIPV10_RBS32_nothing_30h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2_30h.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] | ||
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_nothing_30h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_nothing_30h.jpg|thumb|200px|center|DSMZ15372 with nothing added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_nothing_2_30h.jpg|thumb|200px|center|DSMZ15372 with nothing added in the culture, after 30 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_gly_30h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_gly_30h.jpg|thumb|200px|center|DSMZ15372 with 2% glycerol added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_gly_2_30h.jpg|thumb|200px|center|DSMZ15372 with 2% glycerol added in the culture, after 30 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_IPTG_30h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_IPTG_30h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_IPTG_2_30h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG in the culture, after 30 hours]] |
|- | |- | ||
| - | |[[Image:UNIPV10_PBHR68_gly_IPTG_30h.jpg|thumb|200px|center| | + | |[[Image:UNIPV10_PBHR68_gly_IPTG_30h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG and 2% glycerol added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_gly_IPTG_2_30h.jpg|thumb|200px|center|DSMZ15372 with 1mM IPTG and 2% glycerol added in the culture, after 30 hours]] |
|} | |} | ||
Revision as of 22:23, 27 October 2010
PHB production
Methods
Preparation of samples for BioPlastic (PHB) screening:
- Inoculum in LB+Amp of:
- DSMZ15372 E. coli strain
- <partinfo>BBa_B0032</partinfo>
- Cultures were left grow ON at +37°C, 220 rpm.
- Cultures of DSMZ15372 and <partinfo>BBa_B0032</partinfo> were diluted 1:100 in fresh LB+Amp and than prepared as follows:
- <partinfo>BBa_B0032</partinfo> with NOTHING added (negative control)
- DSMZ15372 with NOTHING added
- DSMZ15372 + 2% glycerol (a carbon source for bioplastic production)
- DSMZ15372 + 1mM IPTG (inducer for lac promoter, expressing bioplastic enzymes)
- DSMZ15372 + 2% glycerol + 1mM IPTG
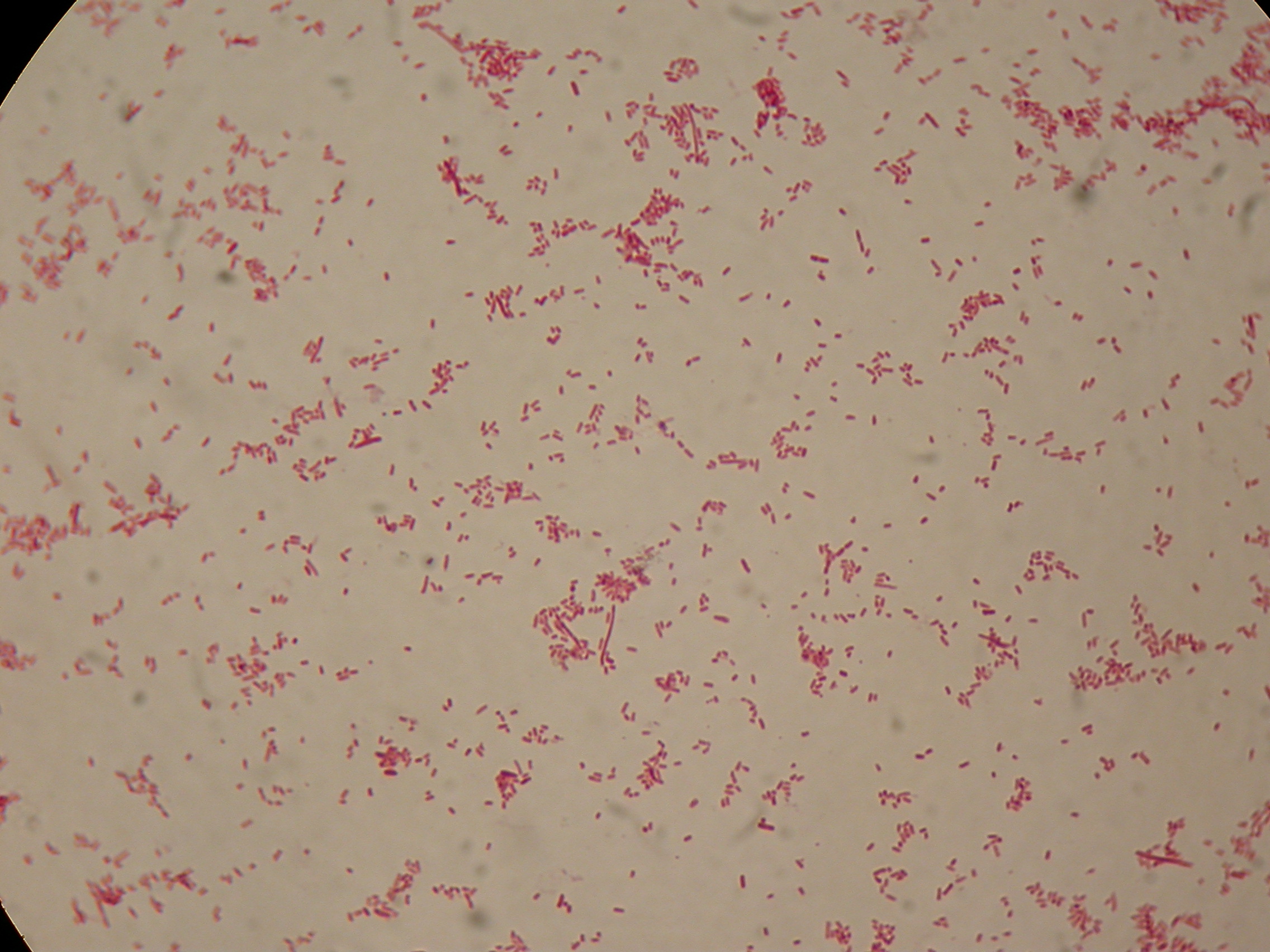
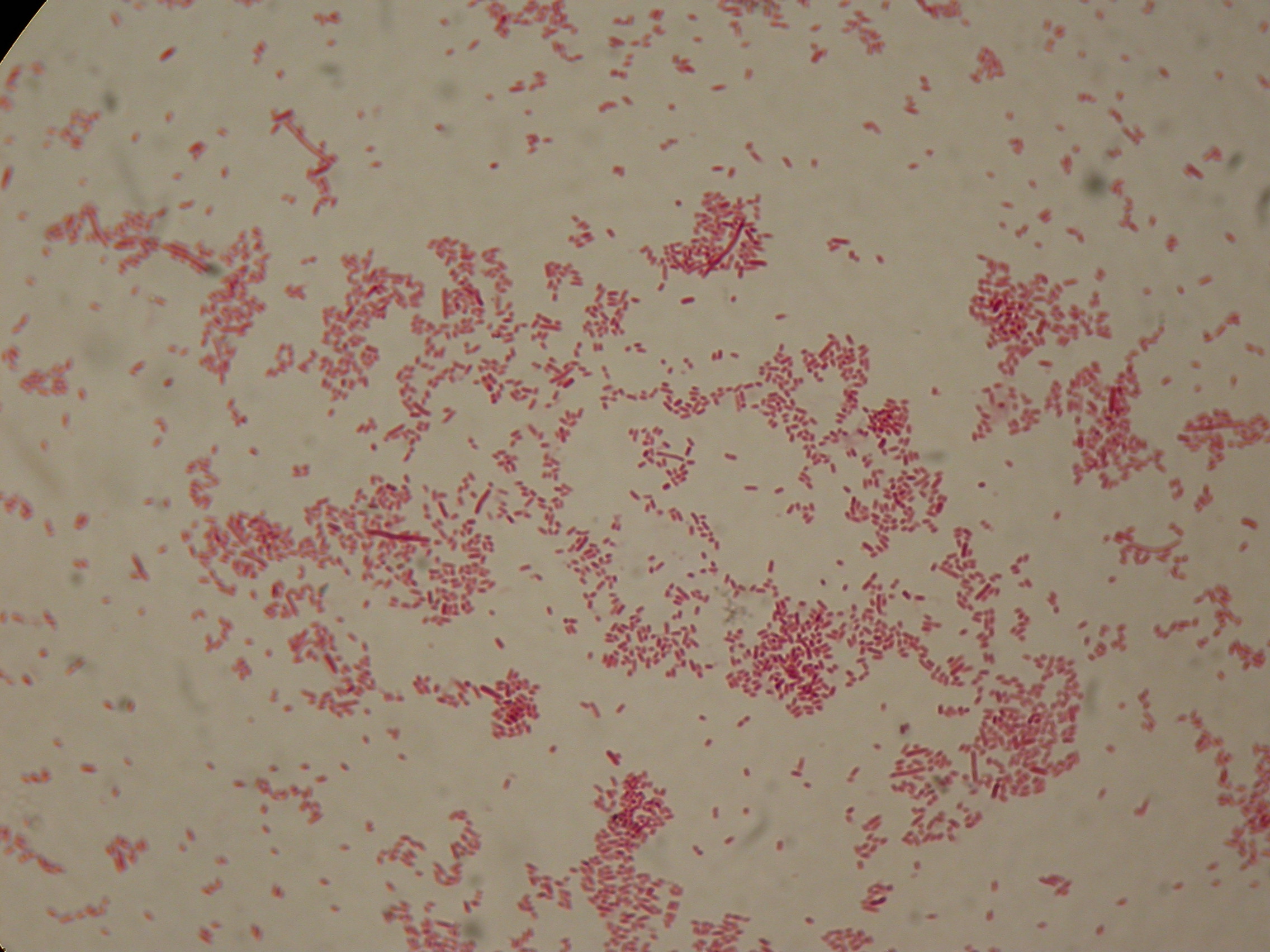
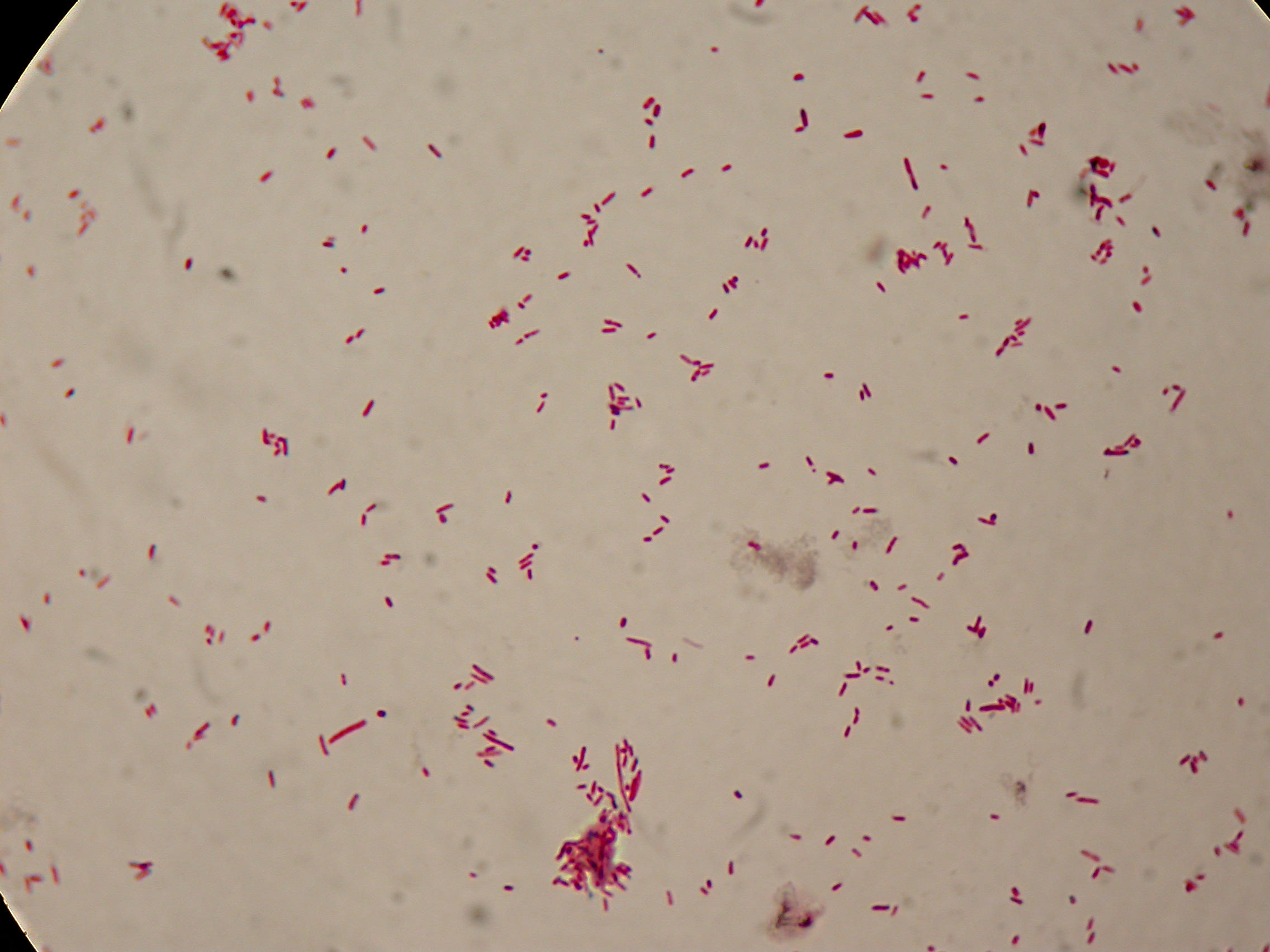
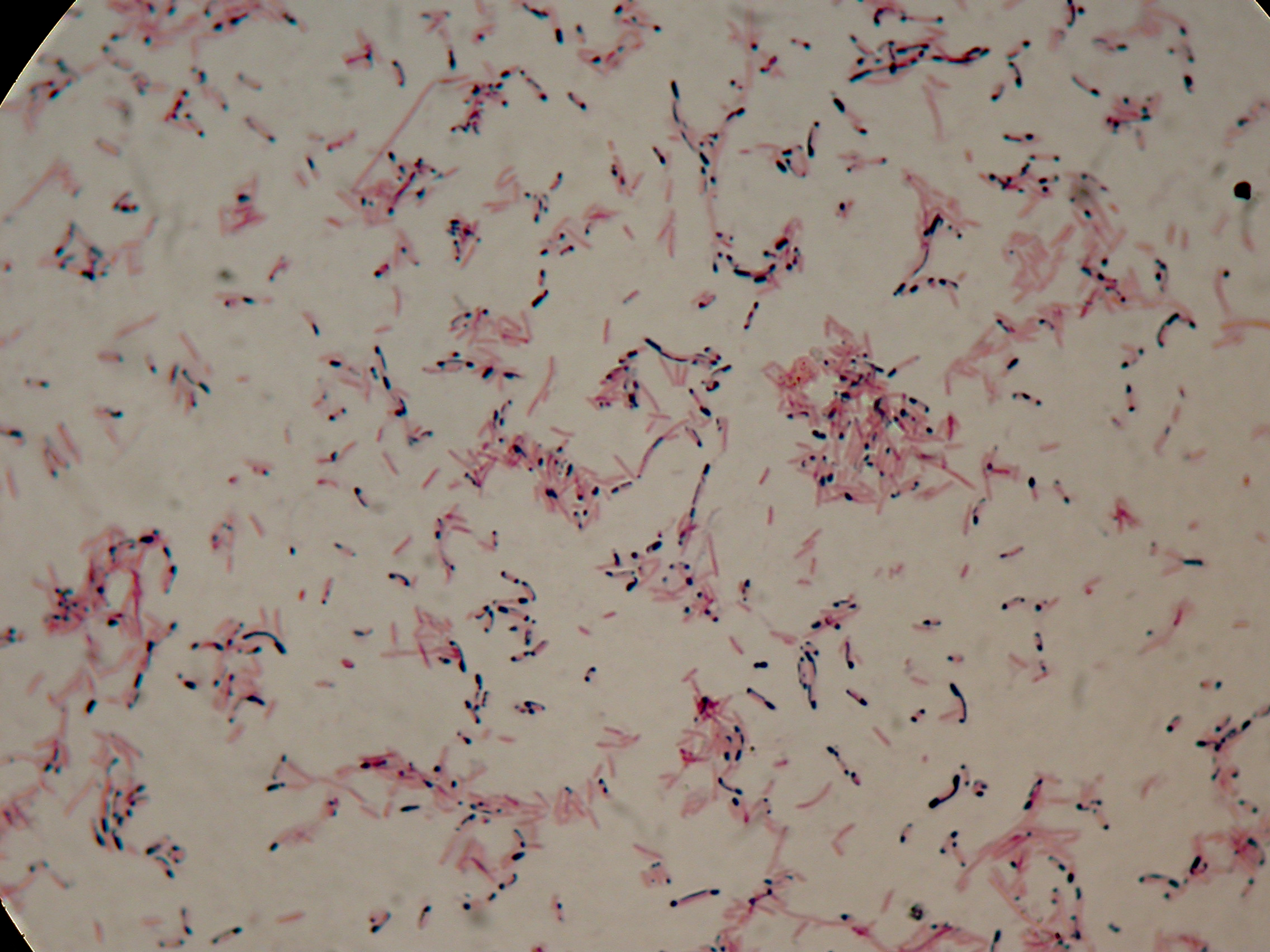
After 5 and 30 hours' growth, Sudan Black staining protocol was performed on slides prepared for each culture. Slides were observed at the microscope.
Results
5 hours'
Sudan Black staining protocol was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here:
30 hours'
Sudan Black staining protocol was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here:
Discussion
In images it's clearly possible to see that after 5 hours PBHR68 without any kind of addition is completely similar to negative control <partinfo>BBa_B0032</partinfo> and there is no trace of bioplastic granules production. In PBHR68 (E. coli cultures harbouring the pBHR68 plasmid) samples in which medium IPTG or glycerol was added, it's possible to see very small dark signs that could be identified as PHB granules in a few bacteria. In samples with addiction of both IPTG and glycerol bioplastic granules are clearly visible in many cells.
As expected after 30 hours negative control <partinfo>BBa_B0032</partinfo> does not show any trace of granules, while PBHR68 show bioplastic granules in each experimental condition. This shows that PHB can be produced both without the presence of the inducer (leakage activity of the plasmid) and glycerol (bacteria found another kind of carbon sources and used it for the synthesis).
This way the ability of E. coli to synthesize PolyHydroxyAlkanoates that can be used in the purification method presented in this project was demonstrated.
Fusion protein synthesis
These experiments were performed to check bacterial growth and GFP synthesis rate of the following constructs (and efficiency of inducible system of some of them) in order to verify right protein folding.
Costitutive promoter devices
Methods
Inoculum (into 5 ml LB+Amp) from glycerol stock of:
- <partinfo>BBa_K300086</partinfo>
- <partinfo>BBa_K300088</partinfo>
- <partinfo>BBa_K300090</partinfo>
- <partinfo>BBa_K300099</partinfo>
- <partinfo>BBa_K173000</partinfo> (positive control)
- <partinfo>BBa_B0031</partinfo> (negative control)
Cultures were grown ON at 37°C, 220 rpm.
The following day cultures were diluted 1:100 and let grow again for about five hours at 37°C, 220 rpm.
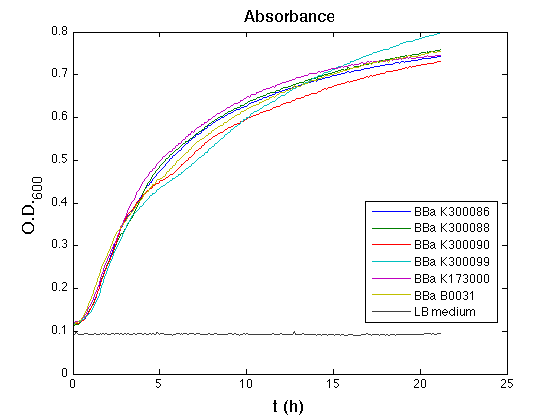
The optical density (O.D.) of each cell culture was than measured with TECAN Infinte F200. Samples were diluted in order to obtain the same O.D. equal to 0,02.
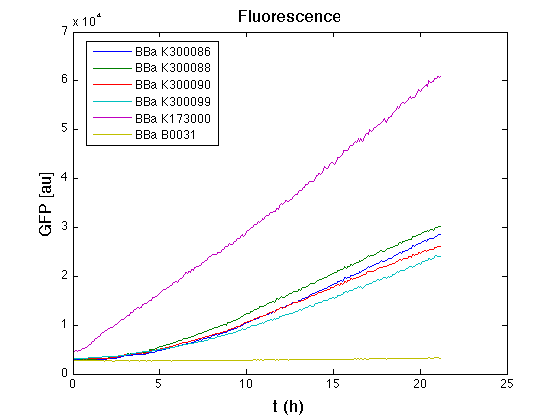
Then we performed a 21-hours' experiment with measurements of absorbance and green fluorescence every five minutes with TECAN Infinite F200; cultures were shaken for 15 seconds every five minutes. Each value shown below is the mean of three measurements, from GFP data that of a non-fluorescent culture (negative control) was subtracted.
Results
| Culture | Doubling time [min.] ± std error |
|---|---|
| <partinfo>BBa_K173000</partinfo> | 76.3336 ± 1.4362 |
| <partinfo>BBa_K300086</partinfo> | 73.6685 ± 1.6245 |
| <partinfo>BBa_K300088</partinfo> | 74.8806 ± 2.7699 |
| <partinfo>BBa_K300090</partinfo> | 75.9433 ± 3.6808 |
| <partinfo>BBa_K300099</partinfo> | 78.4634 ± 2.5622 |
| <partinfo>BBa_B0031</partinfo> | 70.8421 ± 2.2181 |
Discussion
All cell cultures showed a similar growth curve; doubling time was computed as described here in order to have informations about the burden due to the synthesis of such fusion proteins. It's possible to see that all doubling time are very similar; it's possible to assert that the expression of these BioBrick parts doesn't cause abnormal stress to the cells.
From GFP curve it's possible to appreciate that in <partinfo>BBa_K300086</partinfo>, <partinfo>BBa_K300088</partinfo>, <partinfo>BBa_K300090</partinfo>, <partinfo>BBa_K300099</partinfo> GFP accumulation it's very similar and it's significantly different from that of negative control <partinfo>BBa_B0031</partinfo>. These results show that the green fluorescent protein assembled downstream of the genetic circuit is correctly folded.
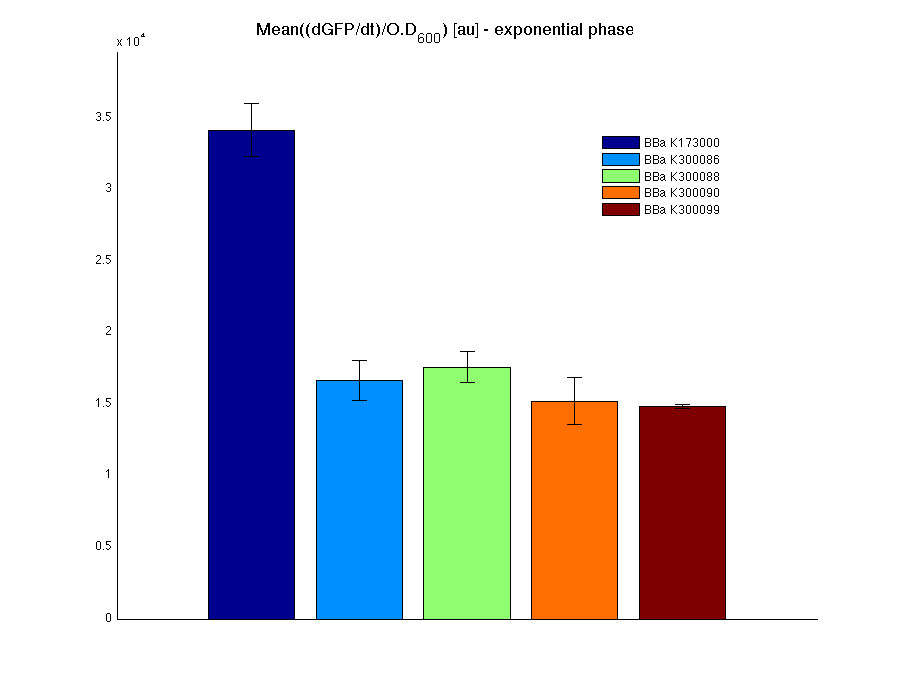
The mean protein synthesis rate was also computed over the growth exponential phase, showing again an appreciable GFP production rate that is about a half of the positive control.
3OC6HSL inducible devices
Methods
Inoculum (into 5 ml LB+Amp) from glycerol stock of:
- <partinfo>BBa_K300091</partinfo>
- <partinfo>BBa_K300092</partinfo>
- <partinfo>BBa_K173000</partinfo> (positive control)
- <partinfo>BBa_B0031</partinfo> (negative control)
Cultures were grown ON at 37°C, 220 rpm.
The following day cultures were diluted 1:100 and let grow again for about five hours at 37°C, 220 rpm.
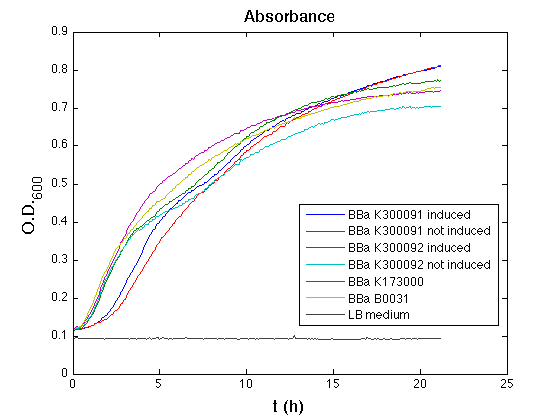
The optical density (O.D.) of each cell culture was than measured with TECAN Infinte F200. Samples were diluted in order to obtain the same O.D. equal to 0,02.
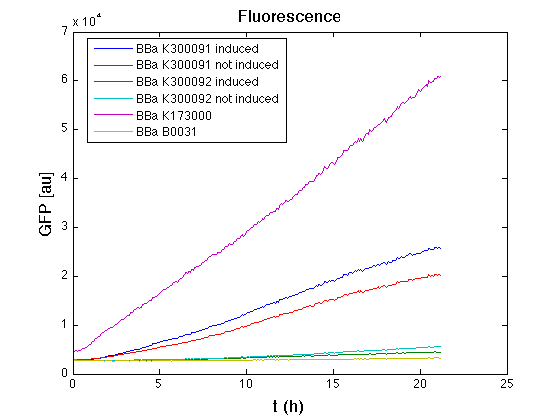
Then we performed a 21-hours' experiment with measurements of absorbance and green fluorescence every five minutes using TECAN Infinite F200; cultures were shaken for 15 seconds every five minutes. <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> circuits were induced 100nM with HSL directly into multiplate well. Each value shown below is the mean of three measurements, from GFP data that of a non-fluorescent culture (negative control) was subtracted.
Results
| Culture | Doubling time [min.] ± std error |
|---|---|
| <partinfo>BBa_K173000</partinfo> | 76.3336 ± 1.4362 |
| <partinfo>BBa_K300091</partinfo> induced | 121.1434 ± 7.0275 |
| <partinfo>BBa_K300091</partinfo> not induced | 74.4267 ± 1.3696 |
| <partinfo>BBa_K300092</partinfo> induced | 122.6088 ± 1.2785 |
| <partinfo>BBa_K300092</partinfo> not induced | 71.5105 ± 2.7113 |
| <partinfo>BBa_B0031</partinfo> | 70.8421 ± 2.2181 |
Discussion
All cell cultures showed a similar growth curve; doubling time was computed as described here in order to have informations about the burden due to the synthesis of such fusion proteins. It's possible to see that all doubling time are very similar except for induced cultures. In this case doubling time is much higher than posite control and not induced cultures; so it's possible to assert that in this case there's a kind of metabolic burden higher than in the others, maybe because of the inducible system.
From GFP curve it's possible to appreciate that in induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> GFP accumulation profile it's very similar and it's significantly different from that of negative control <partinfo>BBa_B0031</partinfo>. On the other hand not induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> show a profile very similar to the last one. These results show that the green fluorescent protein assembled downstream of the genetic circuit is correctly folded and that the inducible system works as expected.
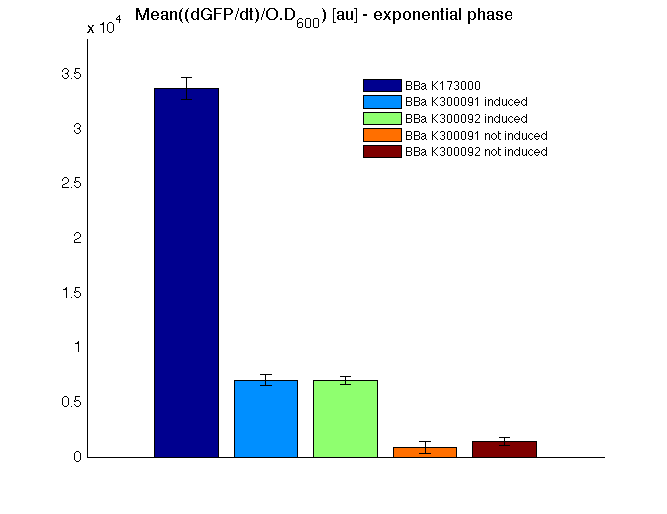
The mean protein synthesis rate was also computed over the growth exponential phase, showing again an GFP production rate that is different from negative control. Not induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> show a low GFP synthesis rate maybe due to 3OC6HSL inducible circuit leakage activity.
 "
"