Team:Wisconsin-Madison/results
From 2010.igem.org
Encapsulation
| Part Number | Function | Expression Type | Zip File |
| <partinfo>BBa_k318500</partinfo> | Produces Trascription Factor RcsB | Inducible - IPTG | 500 |
| <partinfo>BBa_k318501</partinfo> | Produces Trascription Factor RcsA | Inducible - IPTG | 501 |
| <partinfo>BBa_k318502</partinfo> | Produces Trascription Factor RcsA & RcsB | Inducible - IPTG | 502 |
| <partinfo>BBa_k200021</partinfo> | Empty Vector/Contol | Inducible - IPTG | NA |
Our goals is to have each cell be surrounded by a protective 'capsule' to allow them to safely travel through the harsh acidic environment of the stomach to arrive in the small intestine for their main purpose. Imperial 2009 used Transcription Factor RcsB to stimulate a the production of Colonic Acid from the capsule synthesis pathway of E.coli. Colonic Acid s a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Colonic Acid has been shown to increase cell survivability in acidic conditions. RcsA and RcsB are transcription factors that are know to be positive regulators of capsular polysaccharides synthesis. We placed RcsA, RcsB, and a combination of the two under a IPTG inducible promoter to test both quantity of colonic acid produced and cell survivability. These two transcription factors form a heterodimer that is know to activate around 19 genes related to colonic acid synthesis. RcsB is also know to form a homodimmer and positively regulate cell division. RcsA and RcsB belong to the multicomponent RcsF/RcsC/RcsD/RcsA-RcsB phosphorelay system.
Colonic Acid Quantification
Background
Colonic Acid is a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Biological extracts often contain compounds, which under heating with H2SO4 yield brown products absorbing between 396 nm and 427 nm. Colonic acid can be estimated by measuring L-fucose content.
Download procedure here or continue to experimental protocol page
Results
Conclusion
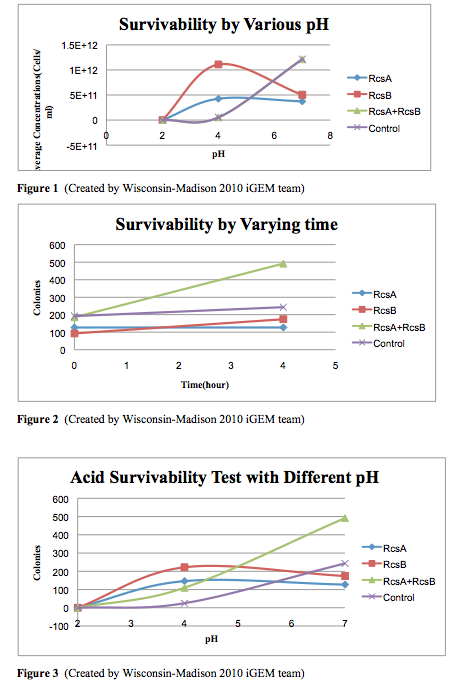
Cell Survivability in Low pH
Background
Download procedure here or continue to experimental protocol page
Results
Conclusion
Timed Lysis
| Part Number | Function | Induction | Zip File |
| <partinfo>BBa_k318513</partinfo> | Produces RFP | stationary phase | 513 |
Background
gadAp: A pH Sensitive Promoter Based on availble literature (see Project Page), we expected an increase in expression from gadAp (BBa_K318512) under both acidic pH conditions (from pH 5 to 3) and in the stationary phase (after an OD of 1 or 2). As most previous studies of gadAp had focused on gadA mRNA production, we wanted to instead explore the viability of gadAp for use in expression of protein. Given the stressful conditions under which it acts and its complex regulation, the possiblity that mRNA-level regulation or an inability of acid-stressed cells to produce large quantities of protein may exist could not be discounted. As such, we combined the gadA promoter with a RFP gene (BBa_K318513) to directly test its ability to produce protein under stationary phase and acid stress conditions.
Low pH Induction
See procedure at experimental protocols page.
Stationary Phase Induction
See procedure at experimental protocols page.
Conclusion
 "
"