Team:Wisconsin-Madison/notebook/Yue
From 2010.igem.org
| (2 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
float: left; | float: left; | ||
position: fixed; | position: fixed; | ||
| - | width: | + | width: 200px; |
margin-left: 0px; | margin-left: 0px; | ||
margin-right: 0px; | margin-right: 0px; | ||
| Line 13: | Line 13: | ||
#page_content { | #page_content { | ||
float: left; | float: left; | ||
| - | width: | + | width: 710px; |
text-align: justify; | text-align: justify; | ||
| - | margin-left: | + | margin-left: 200px; |
margin-right: auto; | margin-right: auto; | ||
padding-left: 0px; | padding-left: 0px; | ||
| Line 27: | Line 27: | ||
<div id="page_content"> | <div id="page_content"> | ||
</html> | </html> | ||
| - | |||
| - | + | =Note book of July 12th,2010= | |
| - | + | 1. Screening for the Rfal into LacI-RBS cloning | |
| - | + | Gel Picture | |
| - | + | [[File:IGEM 2010-07-12 13hr 39min.jpg]] | |
| - | + | [[File:IGEM 2010-07-12 17hr 33min.jpg]] | |
| - | For | + | 2. Primer Design For the Cloning LacI-RBS-ygiV-RBS-Rfal-RBS-RFP-Terminator |
| - | + | One is for Rfal: the forward primer has a XbaI and RBS in it; the reverse primer is on the backbone, and has a XmaI site on it. | |
| - | + | pRBS-Rfal_Fwd: TGCTCTAGA AAAGAGGAGAAAATGCTAACATCCTTTAAACT | |
| - | + | pRBS-Rfal_Rev: CCCCCCGGGTTAATTAATTGTATTGTTACGATTAT | |
| - | + | ||
| - | + | ||
| - | |||
| - | + | The other one is For RFP (I13507): The forward primer has a XmaI site on it and starts from the backbone; the reverse primer starts on the backbone too. [The original plasmid, I13507 has a structur of RBS-RFP-terminator] | |
| - | + | pRFP(I13507)_Fwd: CCCCCCGGGgagaaagaggagaaatactagatgg | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | pRFP(I13507)_Rev: AGTCAGTGAGCGAGGAAG | |
| - | ==== | + | |
| - | + | =Note book of July 13th,2010= | |
| + | |||
| + | 1. Primer Designs: | ||
| + | |||
| + | Tyler helped me check the primers, and he get back to me with his comments: | ||
| + | |||
| + | pRBS-RfaI_Fwd - Tm for primer portion is too low(oligocalc says 44), add 4 bases from the gene sequence, try to have 3' end be G or C | ||
| + | |||
| + | pRBS-RfaI_Rev - GC content of the priming portion is way too low, try to get it to at least above 30%, you can move down the gene since you are at the ending site, also try to have the 3' end of the primer be a G or C | ||
| + | |||
| + | pRFP(I13507)_Fwd - good, I usually add a 5 base overhang, but 3 should be ok for the enzyme you are using | ||
| + | |||
| + | pRFP(I13507)_Fwd - good | ||
| + | |||
| + | So I redesigned my primers with | ||
| + | |||
| + | Detailed information are in: | ||
| + | [[File:Primer Design for Rfal and RFP - 2.pdf]] | ||
| + | |||
| + | '''Primers For Rfal''' | ||
| + | |||
| + | Tm : 46 GC:41% | ||
| + | Fwd: gagatgctaacatcctt (17bp) | ||
| + | |||
| + | pRBS-RfaI_Fwd : TGCTCTAGAAAAGAGGAGAAAtacgagatgctaacatcctt | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C | ||
| + | seq_1 tgctctagaaaagaggagaaatacgagatgctaacatcctt 75.1 39.0 41 16.0 9.0 7.0 9.0 417600.0 12674.4 2.4 30.4 | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 gagatgctaacatcctt 49.9 41.2 17 5.0 5.0 4.0 3.0 164400.0 5169.4 6.1 31.4 | ||
| + | |||
| + | Tm:49 GC:40% | ||
| + | Rev: taataatactagtagcggcc (17bp) | ||
| + | |||
| + | pRBS-RfaI_Rev: GCCGCTACTAGTATTATTA [SpeI] | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 gccgctactagtattatta 49.6 36.8 19 5.0 7.0 4.0 3.0 185500.0 5777.8 5.4 31.1 | ||
| + | |||
| + | '''Primers For RFP''' | ||
| + | |||
| + | pRFP(I13507)_Fwd: CTAGTCTAGAgagaaagaggagaaatactagatgg [XbaI] | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 ctagtctagagagaaagaggagaaatactagatgg 65.7 40.0 35 15.0 6.0 3.0 11.0 377400.0 10950.2 2.6 29.0 | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 gagaaagaggagaaatactagatgg 59.8 40.0 25 12.0 3.0 1.0 9.0 278000.0 7861.2 3.6 28.3 | ||
| + | |||
| + | pRFP(I13507): AGTCAGTGAGCGAGGAAG (cttcctcgctcactgact) | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 agtcagtgagcgaggaag 58.8 55.6 18 6.0 2.0 2.0 8.0 191900.0 5637.7 5.2 29.4 | ||
| + | |||
| + | Name Sequence Tm°C CG% nt A T C G | ||
| + | seq_1 agtcagtgagcgaggaag 58.8 55.6 18 6.0 2.0 2.0 8.0 191900.0 5637.7 5.2 29.4 | ||
| + | |||
| + | =Note book of July 14th,2010= | ||
| + | |||
| + | Cloning of LacI-RBS-ygiV with a terminator | ||
| + | |||
| + | a. Double digestion | ||
| + | |||
| + | b. Gel extraction and purificaiton | ||
| + | |||
| + | [[File:2.jpg]] | ||
| + | |||
| + | c. overnight ligation | ||
| + | |||
| + | =Note book of July 15th,2010= | ||
| + | |||
| + | Cloning of LacI-RBS-Rfal with a terminator | ||
| + | |||
| + | a. Double digestion | ||
| + | |||
| + | b. Gel extraction and purificaiton | ||
| + | |||
| + | [[File:1.jpg]] | ||
| + | |||
| + | c. 2 hours bench top ligation | ||
| + | |||
| + | d. Electroporation of LacI-RBS-ygiV-terminator and LacI-RBS-Rfal-terminator into DH10B cells. | ||
| + | |||
| + | e. Plate on Amp plates | ||
| + | |||
| + | f. run a gel to check the digestion of the backbone | ||
| + | |||
| + | [[File:3.jpg]] | ||
| + | |||
| + | =Note book of July 16th,2010= | ||
| + | |||
| + | A.Check the plate | ||
| + | |||
| + | 1. LacI-RBS-ygiV-terminator | ||
| + | |||
| + | the pallet plate has about 200 colonies | ||
| + | |||
| + | the control has 8 colonies | ||
| + | |||
| + | |||
| + | 2.LacI-RBS-Rfal-terminator | ||
| + | |||
| + | the pallet plate has about 150 colonies | ||
| + | |||
| + | the control doesn't have colonies | ||
| + | |||
| + | |||
| + | B.Screening | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Note book of July 6th,2010= | ||
| + | |||
| + | A. Start the clone of Rfal into K200021(LacI+RBS) | ||
| + | |||
| + | 1.cut the backbone, K200021, with PstI-HF and SpeI for 2 hours. | ||
| + | cut the insert, Rfal, with PstI-HF and XbaI for 2 hours. | ||
| + | |||
| + | 2.Gel Extract | ||
| + | |||
| + | K200021: 3264bp Rfal:1263bp | ||
| + | |||
| + | [[File:IGEM 2010-07-06 13hr 05min.jpg]] | ||
| + | |||
| + | After the gel extraction, the concentrations are: | ||
| + | |||
| + | K200021: 33.1ng/ul | ||
| + | Rfal: 53.9ng/ul | ||
| + | |||
| + | 3.Ligation | ||
| + | |||
| + | for doing a K200021:Rfal is 1:4 ligation, 100ng of K200021 and 154.8ng of Rfal are needed. Based on the DNA concentration, add 3ul K200021 and 3ul Rfal for ligation. | ||
| + | |||
| + | Stored at 16C overnight. | ||
| + | |||
| + | |||
| + | =Note book of July 7th,2010= | ||
| + | |||
| + | A. Cloning of LacI-RBS-Rfal | ||
| + | |||
| + | 1.Electroporation | ||
| + | |||
| + | LacI-RBS-Rfal has a time constant 4.8; | ||
| + | |||
| + | The control has a time constant 4.7; | ||
| + | |||
| + | 2.Plate on two Amp+Kan plate. Stored in a 37C incubator. | ||
| + | |||
| + | B. Cloning of LacI-RBS-ygiV into RBS | ||
| + | |||
| + | 1.cut the backbone, RBS, with EcoRI-HF and XbaI, and cut the insert LacI-RBS-ygiV with EcoRI-HF and SpeI. | ||
| + | |||
| + | 2.Gel Extraction | ||
| + | |||
| + | RBS:3200bp LacI-RBS-ygiV:543bp | ||
| + | |||
| + | [[File:IGEM 2010-07-06 17hr 00min.jpg]] | ||
| + | |||
| + | after the gel extraction, the concentrations are: | ||
| + | |||
| + | RBS: 20.7ng/ul | ||
| + | |||
| + | lacI-RBS-ygiV: 19.8ng/ul | ||
| + | |||
| + | 3.Ligation | ||
| + | |||
| + | For a 1 to 3 backbone:insert ratio, we need 100ng of RBS and 107.3ng of LacI-RBS-ygiV for Ligation. THerefore, add 3.7ul RBS and 4.3ul LacI-RBS-ygiV to ligate. Store at 4C overnight. | ||
| + | |||
| + | C.Run a gel to check the cutting peices | ||
| + | |||
| + | The orders are: | ||
| + | |||
| + | L-ygiV-ES; L-ygiV uncut; Rfal-PX; Rfal uncut; RBS-EX; RBS uncut; LacI-PS; LacI uncut; Ladder | ||
| + | |||
| + | unluckily, the first lane for L-ygiV-ES didn't show up any band. | ||
| + | |||
| + | [[File:IGEM 2010-07-07 11hr 24min.jpg]] | ||
| + | |||
| + | The rest of the bands are all correct. | ||
| + | |||
| + | Then run a gel for L-ygiV-ES and L-ygiV uncut by loading more DNA on the gel | ||
| + | |||
| + | [[File:IGEM 2010-07-07 13hr 09min.jpg]] | ||
| + | |||
| + | Now, this shows that the L-ygiV-ES is correct. | ||
| + | |||
| + | |||
| + | =Note book of July 8th,2010= | ||
| + | |||
| + | A. Transformation of the Clone LacI-RBS-ygiV-RBS | ||
| + | |||
| + | 1. Electroporation | ||
| + | |||
| + | LacI-RBS-ygiV-RBS: 4.7 | ||
| + | |||
| + | Control: 4.9 | ||
| + | |||
| + | 2. Plate on two Amp plate. | ||
| + | |||
| + | B. Run a gel to check the Ligation of LacI-RBS-Rfal and LacI-RBS-ygiV-RBS | ||
| + | |||
| + | the orders are Ladder LacI-RBS-Rfal, control, LacI-RBS-ygiV-RBS, control | ||
| + | |||
| + | [[File:IGEM 2010-07-07 12hr 48min.jpg]] | ||
| + | |||
| + | C. Colony PCR for the LacI-RBS-Rfal Clone | ||
| + | |||
| + | 1.The control plate only has one colony, compared with the regular plate with about 100 colonies. | ||
| + | |||
| + | 2.picked 14 colonies for screening. | ||
| + | |||
| + | Gel for Colony PCR: | ||
| + | |||
| + | [[File:IGEM 2010-07-08 13hr 54min.jpg]] | ||
| + | |||
| + | Run for 5mins longer and add more EB | ||
| + | |||
| + | [[File:IGEM 2010-07-08 14hr 20min.jpg]] | ||
| + | |||
| + | No colony shows the correct band (around 1500bp) | ||
| + | |||
| + | =Note book of July 9th,2010= | ||
| + | |||
| + | A Cloning of LacI-RBS-ygiV into RBS, and Rfal into LacI-RBS. | ||
| + | |||
| + | Add 1.5mg of DNA into each tube: | ||
| + | |||
| + | RBS(B0034):5.3ul template; Add 1ul EcoRI-HF to digest 1 hour and inactivate at 65C for 20mins. Then Add 1ul of XbaI to digest an additional hour. | ||
| + | |||
| + | LacI-RBS(K200021): 9.5ul template; Add 1ul PstI-HF to digest 1 hour and inactivate at 80C for 20mins. Then Add 1ul of SpeI to digest an additional hour. | ||
| + | |||
| + | Rfal: 3.1ul template digest with 1ul PstI-HF and 1ul XbaI | ||
| + | |||
| + | LacI-ygiV: 4.4ul tempalte digest with 1ul EcoRI-HF and 1ul SpeI | ||
| + | |||
| + | Gel after 1 hour inactivation of EcoRI-HF in RBS and PstI-HF in LacI-RBS: | ||
| + | |||
| + | Orders are: | ||
| + | |||
| + | RBS cut with EcoRI-HF, RBS uncut, Ladder, LacI-RBS cut with PstI-HF, LacI-RBS uncut. | ||
| + | |||
| + | [[File:IGEM 2010-07-09 13hr 27min.jpg]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Note book of June 28th,2010= | ||
| + | |||
| + | a. Digestion of K200021 (LacI+RBS) with PstI-HF and SpeI, and digest ygiV and Rfal with PstI-HF and XbaI. | ||
| + | |||
| + | b. Gel Extraction and purification: | ||
| + | |||
| + | 1. Gel | ||
| + | |||
| + | K200021-PS | ||
| + | |||
| + | ygiV and Rfal | ||
| + | |||
| + | [[File:IGEM 2010-06-27 13hr 19min.jpg]] | ||
| + | |||
| + | Concentration of the cutting peices: | ||
| + | K200021-PS | ||
| + | |||
| + | ygiV-PX | ||
| + | |||
| + | Rfal-PX | ||
| + | |||
| + | Gel for the purification product | ||
| + | |||
| + | [[File:IGEM 2010-06-28 14hr 10min.jpg]] | ||
| + | |||
| + | c. Set up an overnight ligation | ||
| + | |||
| + | =Note book of June 29th,2010= | ||
| + | |||
| + | a. Transformation | ||
| + | |||
| + | pLacI-RBS-ygiV-PX: 4.1 | ||
| + | |||
| + | pLacI-RBS-ygiV-PX(C): 3.9 | ||
| + | |||
| + | pLacI-RBS-Rfal-PX: 4.0 | ||
| + | |||
| + | pLacI-RBS-Rfal-PX(C):3.9 | ||
| + | |||
| + | b. Plate on Amp100 plates. | ||
| + | |||
| + | |||
| + | =Note book of June 30th,2010= | ||
| + | |||
| + | Re-plan the project. | ||
| + | Figure out all the cloning need to be done by the end of the summer. | ||
| + | Detailed information please see the spread sheet on Sarah's page. | ||
| + | |||
| + | =Note book of July 1st,2010= | ||
| + | |||
| + | A. Colony PCR of the LacI-RBS-ygiV and LacI-RBS-Rfal clone. | ||
| + | |||
| + | Upper: LacI-RBS-Rfal Clone | ||
| + | |||
| + | Lower: LacI-RBS-ygiV clone | ||
| + | |||
| + | [[File:IGEM 2010-07-01 14hr 49min-2.jpg]] | ||
| + | |||
| + | The LacI-RBS-ygiV clone shows up at the right length. | ||
| + | |||
| + | |||
| + | B. Innoculate 5 liquid culture of the ygiV clone. | ||
| + | |||
| + | |||
| + | =Note book of July 2nd,2010= | ||
| + | |||
| + | A. Made 3 freezer stocks of LacI-RBS-ygiV clone | ||
| + | |||
| + | B. Miniprep LacI-RBS-ygiV | ||
| + | |||
| + | LacI-RBS-ygiV-1: 388.7ng/ul | ||
| + | LacI-RBS-ygiV-2: 357.0ng/ul | ||
| + | LacI-RBS-ygiV-3: 350.2ng/ul | ||
| + | LacI-RBS-ygiV-4: 255.0ng/ul | ||
| + | |||
| + | |||
| + | =Note book of June 21st,2010= | ||
| + | |||
| + | a.Minipreped 3 sets each for pBAD33BB, pBAD35BB, ygiV, and Rfal. | ||
| + | |||
| + | b.Set up a overnight digestions that cut pBAD33BB and pBAD35BB with PstI-HF and SpeI, and cut ygiV and Rfal with PstI-HF and XbaI. (37C overnight) | ||
| + | |||
| + | c.Screening for the 2nd time ygiV and Rfal cloning. | ||
| + | |||
| + | ygiV: | ||
| + | |||
| + | [[File:IGEM 2010-06-21 bads2 Peter ScanA.jpg]] | ||
| + | |||
| + | Rfal: | ||
| + | |||
| + | [[File:IGEM 2010-06-21 bads2 Peter ScanB.jpg]] | ||
| + | |||
| + | They all shows up at the same length as the positive control, which means we didn't get the desired cloning. | ||
| + | |||
| + | =Note book of June 22nd,2010= | ||
| + | |||
| + | a.Gel extraction of pBAD33BB, ygiV and Rfal. | ||
| + | |||
| + | For pBAD33BB | ||
| + | |||
| + | [[File:IGEM 2010-06-23 12hr 45min.jpg]] | ||
| + | |||
| + | For ygiV(486bp) and Rfal(1269bp) | ||
| + | |||
| + | [[File:IGEM 2010-06-23 12hr 44min.jpg]] | ||
| + | |||
| + | b.Gel purification of pBAD33BB, ygiV and Rfal. | ||
| + | |||
| + | after the gel purificaion, the concentrations are: | ||
| + | |||
| + | pBAD33BB-PX: 39.7ng/ul | ||
| + | |||
| + | ygiV: 25.7ng/ul | ||
| + | |||
| + | Rfal: 33.5ng/ul | ||
| + | |||
| + | c.Set up an overnight ligation with Insert/Vector ratio equals 4. | ||
| + | |||
| + | tubeA | ||
| + | |||
| + | pBAD33BB: 3.8ul | ||
| + | |||
| + | ygiV: 2.2ul | ||
| + | |||
| + | |||
| + | tubeB | ||
| + | |||
| + | pBAD33BB: 3.8ul | ||
| + | |||
| + | Rfal: 4.2ul | ||
| + | |||
| + | |||
| + | tube C (control) | ||
| + | |||
| + | pBAD33BB: 3.8ul | ||
| + | |||
| + | (16C overnight) | ||
| + | |||
| + | =Note book of June 23rd,2010= | ||
| + | |||
| + | a. Transformation | ||
| + | |||
| + | pBAD33BB-ygiV: 4.4 1.8 | ||
| + | |||
| + | pBAD33BB-Rfal: 4.4 1.8 | ||
| + | |||
| + | Control: 4.3 1.8 | ||
| + | |||
| + | b. Plating on CM plate | ||
| + | |||
| + | c. run the gel for ligation product | ||
| + | |||
| + | [[File:Pflegerlab 2010-06-24 11hr 07min.jpg]] | ||
| + | |||
| + | |||
| + | =Note book of June 24th,2010= | ||
| + | |||
| + | a. Check the transformation plate | ||
| + | |||
| + | pBAD33BB-ygiV: | ||
| + | |||
| + | [[File:Photo.JPG]] | ||
| + | |||
| + | |||
| + | pBAD33BB-Rfal: | ||
| + | |||
| + | [[File:Photo2.JPG]] | ||
| + | |||
| + | |||
| + | Control: | ||
| + | |||
| + | [[File:Photo1.JPG]] | ||
| + | |||
| + | b. pick 12 colonies from pBAD33BB-Rfal plate and 10 colonies from pBAD33BB-ygiV plate for the colony PCR screening. | ||
| + | |||
| + | ygiV-plate B | ||
| + | |||
| + | [[File:Plate B of ygiV-pBAD33BB screening pflegerlab 2010-06-25 14hr 45min.jpg]] | ||
| + | |||
| + | |||
| + | Rfal-plate B | ||
| + | |||
| + | [[File:Plate A of Rfal-pBAD33BB screening pflegerlab 2010-06-25 14hr 42min.jpg]] | ||
| + | |||
| + | c. pick colonies from both plates into liquid cultures. Grow overnight. (5 from plateA and 5 from plateB) | ||
| - | |||
| - | :: | + | =Note book of June 25th,2010= |
| - | + | ||
| - | + | a. miniprep 10 liquid culture for Screening. | |
| + | |||
| + | b. Digest with BamHI-HF and PstI-HF | ||
| + | |||
| + | c. run the gel for the screening | ||
| + | |||
| + | [[File:Screening.jpg]] | ||
| + | |||
| + | |||
| + | |||
| + | =Summary of Week June 14th to June 18th, 2010= | ||
| + | |||
| + | 06-14-10 | ||
| + | '''A. Miniprep''' | ||
| + | |||
| + | Miniprep All the new parts received | ||
| + | |||
| + | 1. K082006 52.7ng/ul | ||
| + | |||
| + | 2. K142001 196.7ng/ul | ||
| + | |||
| + | 3. K173004 330.2ng/ul | ||
| + | |||
| + | 4. pBAD35BB 102.4ng/ul | ||
| + | |||
| + | 5. pBAD33BB 61.5ng/ul | ||
| + | |||
| + | 6. pBAD35BB 105.8ng/ul | ||
| + | |||
| + | 7. K142003 212.8ng/ul | ||
| + | |||
| + | 8. K137113 174.8ng/ul | ||
| + | |||
| + | 9. K142002 222.5ng/ul | ||
| + | |||
| + | 10.pBAD35BB 117.2ng/ul | ||
| + | |||
| + | 11.K200003 207.2ng/ul | ||
| + | |||
| + | 12.K142000 177.4ng/ul | ||
| + | |||
| + | |||
| + | '''B. Ligation for vgiV and pBAD33BB''' | ||
| + | |||
| + | Ligation of vgiV and pBAD33BB with a molar ratio 3:1 | ||
| + | |||
| + | one for 2 hours, one for overnight | ||
| + | |||
| + | |||
| + | '''C. Thansformation of the vgiV cloning''' | ||
| + | |||
| + | Transformed the 2-hour ligation and plated | ||
| + | |||
| + | 06-15-10 | ||
| + | '''A. Screening of the cloning of vgiV into pBAD33BB''' | ||
| + | |||
| + | The 2-hour ligation only had two colonies. | ||
| + | Screening with Colony PCR but didn't see any band. | ||
| + | |||
| + | '''B. Transformation of vgiV cloning''' | ||
| + | |||
| + | Transformed the overnight ligation and plated in CM argar plate. | ||
| + | |||
| + | '''C. Restriction Mapping of the Newly Receiving Parts''' | ||
| + | |||
| + | The Correct Parts are | ||
| + | |||
| + | Parts Band 1@ Band2@ Band3@ | ||
| + | |||
| + | K112808 2500 1406 | ||
| + | |||
| + | K137113 2200 650 | ||
| + | |||
| + | K173004 3200 2100 1106 | ||
| + | |||
| + | K142000 2100 1100 | ||
| + | |||
| + | K142001 2100 1100 | ||
| + | |||
| + | K142002 2100 1100 | ||
| + | |||
| + | K142003 2100 1100 | ||
| + | |||
| + | K082006 3200 750 | ||
| + | |||
| + | K200003 3200 1300 | ||
| + | |||
| + | |||
| + | 06-16-10 | ||
| + | '''A. Screening of the cloning of vgiV into pBAD33BB''' | ||
| + | |||
| + | Saw a lot colony on both the 100ul plate and the pallet plate. The Control plate (backbone only) only had one colony. | ||
| + | |||
| + | Pick up 10 colonies from each plate( 100ul plate and pallet plate). Colony PCR for the screening | ||
| + | |||
| + | Run the Gel for the Coloy PCR, but didn't see the correct band. | ||
| + | |||
| + | (The band is about 200bp, which means the back bone ligate itself.) | ||
| + | |||
| + | '''B. Cloning of Rfal into pBAD35BB''' | ||
| + | |||
| + | Digest pBAD35BB with PstI-HF and XbaI, and digest Rfal with PstI-HF and SpeI | ||
| + | |||
| + | Gel extraction and purification. | ||
| + | |||
| + | Ligate with a insert/vector ratio 3:1 | ||
| + | |||
| + | Ligate at 16C overnight | ||
| + | |||
| + | 06-17-10 | ||
| + | |||
| + | '''A. Another Screening of the cloning of vgiV into pBAD33BB''' | ||
| + | |||
| + | Pick another 10 colonies from both the 100ul plate and the pallet plate. | ||
| + | |||
| + | Proceed the Colony PCR | ||
| + | |||
| + | Run the Gel, but still didn't see the correct band(about 700bp). | ||
| + | |||
| + | Do see a band down at 200bp, same length as the positive control. Therefore, confirmed that the colonies are just transformed with the backbone. | ||
| + | |||
| + | |||
| + | '''B. Cloning of Rfal into pBAD35BB''' | ||
| + | |||
| + | Transform the Rfal cloning and the control into DH10B | ||
| + | |||
| + | Plated on Kan plate. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Cloning of pET28b+RFP | ||
| + | [[File:Cloning of pET28b-RFP.pdf]] | ||
| + | |||
| + | =Notebook May 25th, 2010= | ||
| + | |||
| + | '''BioBrick pBAD35''' | ||
| + | a) Target Vector: pBAD35 (about 4200bp, 4.2kb) | ||
| + | |||
| + | pBAD BioBrick Primers: | ||
| + | Tm value: Fwd: 72.0C | ||
| + | Rev: 68.1C | ||
| + | |||
| + | '''b) General Procedure''' | ||
| + | 1. PCR amplify | ||
| + | 2. Pour 2 gels during this time | ||
| + | 3. Check PCR on 1st Gel | ||
| + | 4. Add 1ul DpnI/rxn for 1hr at 37C | ||
| + | 5. PCR clean up or gel extraction (depends on whether you see a bright band at right place) | ||
| + | 6. XbarI Digest | ||
| + | 7. PCR clean up | ||
| + | 8. Ligate | ||
| + | |||
| + | '''c) Cycle Design''' | ||
| + | 98C 1min | ||
| + | 98C 15s | ||
| + | 73C 30s 33X | ||
| + | 72c 126S | ||
| + | 72c 5mins | ||
| + | 4c forever | ||
| + | |||
| + | '''d)Phusion Mix''' | ||
| + | Water: 35ul | ||
| + | 5Xbuffer:10ul | ||
| + | template:2ul | ||
| + | Primer FW:0.5ul | ||
| + | Primer RV:0.5ul | ||
| + | dNTP:1ul | ||
| + | Phusion Enzyme:1ul | ||
| + | |||
| + | Note: Keep the primers and Enzyme on ICE | ||
| + | |||
| + | '''RESULT''' | ||
| + | 1st Gel | ||
| + | Lane Band@ | ||
| + | 35A 3-4kb | ||
| + | 35B 3-4kb | ||
| + | 18 4kb | ||
| + | 33 5kb | ||
| + | 34 4kb and 2.5kb(wrong) | ||
| + | |||
| + | [[File:IGEM Biobrick first 2010-05-25 15hr 09min.jpg]] | ||
| + | |||
| + | ''' | ||
| + | Protocol of PCR Purification:''' | ||
| + | [[File:PCR purification-Qiagen.pdf]] | ||
| + | |||
| + | '''DIGESTION''' | ||
| + | |||
| + | '''pBAD35-A''' | ||
| + | DNA: 40ul | ||
| + | Buffer4: 5ul | ||
| + | BSA: 0.5ul | ||
| + | Water: 3.5ul | ||
| + | XbaI: 1ul | ||
| + | |||
| + | '''pBAD35-B''' | ||
| + | DNA: 40ul | ||
| + | Buffer4: 5ul | ||
| + | BSA: 0.5ul | ||
| + | Water: 2.5ul | ||
| + | XbaI: 2ul | ||
| + | |||
| + | Leave the digestion on a 37c water bath overnight | ||
| + | |||
| + | ''' | ||
| + | Inoculation for Encryption Project''' | ||
| + | |||
| + | 10ml LB in each tube and add 10ul Antibiotic | ||
| + | A: BBa_K118004 | ||
| + | B: BBa_K098995 | ||
| + | C: BBa_I718008 | ||
| + | D: BBa_B1006 | ||
| + | |||
| + | =Notebook May 26th, 2010= | ||
| + | a)PCR purification(refer to protocol on the protocol from Notebook May 25th 2010) | ||
| + | DNA Conc(ng/ul) | ||
| + | pBAD35-A 57.6 | ||
| + | pBAD35-B 69.7 | ||
| + | |||
| + | b)Ligation | ||
| + | (Peter did all the ligation) | ||
| + | Ligate on pBAD18,33,34,35A-B 100ng/10ul rxn | ||
| + | 2hrs bench ligation | ||
| + | |||
| + | c)Electroporation transformation | ||
| + | |||
| + | Use DH10B | ||
| + | Add 1ul of plasmid | ||
| + | Let it sits on ice for 5 mins | ||
| - | + | after the shock, immediately add 950ul LB and mix well | |
| - | + | Transfer to centrifuge tube | |
| - | + | DNA Time | |
| - | + | 35 4.2 | |
| - | + | 33 4.4 | |
| - | + | 18 4.2 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | (control) | |
| - | + | 35 4.5 | |
| - | + | 33 4.2 | |
| - | + | 18 4.2 | |
| - | + | ||
| - | + | ||
| - | + | Let them grow in the 37c shaker for 1hr. | |
| - | + | ||
| - | + | ||
| - | + | Plate 50ul of cells | |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | '''Miniprep for Nate Pant's Project | ||
| + | Miniprep''' | ||
| + | Plasmid information | ||
| + | A: BBa_K118004 | ||
| + | B: BBa_K098995 | ||
| + | C: BBa_I718008 | ||
| + | D: BBa_B1006 | ||
| - | + | Centrifuge at 30000rpm for 10mins | |
| - | + | Follow the Miniprep Protocol: | |
| + | [[File:Qiagenmini.pdf]] | ||
| + | ''' | ||
| + | Result''' | ||
| + | Measured by Nanodrop | ||
| - | + | DNA Conc(ng/ul) 260/280 | |
| + | A 324.0 1.97 | ||
| + | B 288.3 1.98 | ||
| + | C 418.2 1.96 | ||
| + | D 276.6 1.97 | ||
| - | |||
| - | + | =Notebook May 27th, 2010= | |
| - | + | Check the colonies in the plates. | |
| - | + | Picked 10 colonies from pBAD35B | |
| - | + | Picked 5 colonies from pBAD33 | |
| - | + | Picked 5 colonies from pBAD18 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Inoculate them in 5ml LB with antibiotics | |
| - | + | ''' | |
| + | Parts Checking | ||
| + | ''' | ||
| - | + | Concentration: | |
| - | + | BBa_K200000: 199.9ng/ul | |
| + | BBa_I716213: 186.5ng/ul | ||
| + | BBa_S04114: 543.6ng/ul | ||
| + | BBa_I718007: 146.1ng/ul | ||
| + | BBa_I716212: 155.8ng/ul | ||
| + | BBa_S03975: | ||
| + | BBa_K200002: 309.3ng/ul | ||
| + | BBa_K200003: 105.4ng/ul | ||
| + | BBa_S09373: 243.1ng/ul | ||
| + | BBa_J31000: 278.3ng/ul | ||
| + | BBa_B1006: 276.6ng/ul | ||
| + | BBa_K118004: 324.0ng/ul | ||
| + | BBa_K098995: 288.3ng/ul | ||
| - | + | Single Digestion Test With EcoRI | |
| + | for | ||
| + | 1. BBa_K200000: | ||
| + | 2. BBa_I716213: | ||
| + | 3. BBa_S04114: | ||
| + | 4. BBa_I718007: | ||
| + | 5. BBa_I716212: | ||
| + | 6. BBa_S03975:(Don't have plasmid) | ||
| + | 7. BBa_K200002: | ||
| + | 8. BBa_K200003: | ||
| + | 9. BBa_S09373: | ||
| + | 10.BBa_J31000: | ||
| + | 11.BBa_B1006: | ||
| + | 12.BBa_K118004: | ||
| + | 13.BBa_K098995: | ||
| - | + | Protocol | |
| + | 1.8ul Buffer3(10X) | ||
| + | 0.18ul BSA(100X) | ||
| + | 12ul MiliQ H2O | ||
| + | 3ul DNA | ||
| + | 1ul EcoRI | ||
| - | + | Incubate in water bath at 37C for 2 hours (total volume 18ul) | |
| - | + | add 3.6ul Dye (6X) | |
| - | + | Load 6ul into the GEL | |
| + | Gel: | ||
| + | [[File:2010-05-27 Map - Single digest.jpg]] | ||
| - | + | Gel Description: | |
| - | + | 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 12, 13 | |
| - | + | Results: | |
| + | 3, 9, 11, and 12 don't have the correct bands | ||
| - | + | So run the single digestion for all four again with 2 hours | |
| + | Gel: | ||
| + | [[File:IGEM Single Digestion Second 2010-05-28 14hr 39min.jpg]] | ||
| + | Gel Description: | ||
| + | 3, 9, Ladder, 11, 12 | ||
| - | + | Result: | |
| - | + | Still not correct | |
| - | + | ||
| + | Double Digestion | ||
| + | Find one enzyme cutting site with the gene | ||
| + | 1. BBa_K200000: | ||
| + | Buffer:4 | ||
| + | Enzyme:AflII EcoRI | ||
| + | 2. BBa_I716213: | ||
| + | Buffer:2 | ||
| + | Enzyme:AflIII SpeI | ||
| + | 3. BBa_S04114: | ||
| + | Buffer:3 | ||
| + | Enzyme:EcoRV EcoRI | ||
| + | 4. BBa_I718007: | ||
| + | Buffer:3 | ||
| + | Enzyme:EcoRV EcoRI | ||
| + | 5. BBa_I716212: | ||
| + | Buffer:3 | ||
| + | Enzyme:EcoRV EcoRI | ||
| + | 6. BBa_S03975:(Don't have plasmid) | ||
| + | 7. BBa_K200002: (don't have enzyme stock that can cut within the gene) | ||
| + | 8. BBa_K200003: | ||
| + | Buffer:4 | ||
| + | Enzyme:NheI EcoRI | ||
| + | 9. BBa_S09373: | ||
| + | Buffer:3 | ||
| + | Enzyme:EcoRV EcoRI | ||
| + | 10.BBa_J31000: | ||
| + | Buffer:3 | ||
| + | Enzyme:BglI EcoRI | ||
| + | 11.BBa_B1006: (Too small, cannot cut within the gene) | ||
| + | 12.BBa_K118004: | ||
| + | Buffer:3 | ||
| + | Enzyme:EcoRV EcoRI | ||
| + | 13.BBa_K098995: | ||
| + | Buffer:2 | ||
| + | Enzyme:HindIII EcoRI | ||
| - | + | Protocol | |
| - | + | 1.8ul NEB Buffer(10X) | |
| - | + | 0.18ul BSA(100X) | |
| + | 11ul MiliQ H2O | ||
| + | 3ul DNA | ||
| + | 1ul Enzyme1 | ||
| + | 1ul Enzyme2 | ||
| - | + | Incubate in water bath at 37C for 2 hours (total volume 18ul) | |
| + | add 3.6ul Dye (6X) | ||
| - | + | Load 6ul into the GEL | |
| - | + | Gel | |
| + | [[File:IGEM Parts Double Digestion 2010-05-28 14hr 54min.jpg]] | ||
| + | [[File:IGEM Parts Double Digestion White 2010-05-28 14hr 57min.jpg]] | ||
| - | + | Result | |
| + | Only 1, 2, 4, 5, 8, 13 show the correct bands | ||
| - | |||
| - | == | + | =Notebook May 28th, 2010= |
| - | + | ||
| - | + | Miniprep for the screening | |
| - | + | Restriction Enzyme Mapping for all the 20 screening plasmid | |
| - | + | 1.8ul Buffer3(10X) | |
| + | 0.18ul BSA(100X) | ||
| + | 12ul MiliQ H2O | ||
| + | 3ul DNA | ||
| + | 1ul SpeI | ||
| - | + | Incubate in water bath at 37C for 2 hours (total volume 18ul) | |
| - | + | add 3.6ul Dye (6X) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Load 6ul into the GEL | |
| + | Run the GEL at 100V for 30mins | ||
| + | [[File:IGEM Biobrick 2010-05-28 16hr 55min.jpg]] | ||
| + | GEL upper | ||
| + | 18-1 18-2 18-3 18-4 18-5 Ladder 33-1 33-2 33-3 33-4 33-5 | ||
| - | + | Gel Lower | |
| + | 35-1 35-2 35-3 35-4 35-5 Ladder 35-6 35-7 35-8 35-9 35-10 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | =Notebook June 1st, 2010= | |
| - | |||
| - | + | Search the Detailed info of the Parts we received | |
| + | 1. BBa_K200000: Colanic acid global regulator (RcsB) | ||
| + | 2. BBa_I716213: Cre(TTG Start) | ||
| + | 3. BBa_S04114: Lysis | ||
| + | 4. BBa_I718007: RBS-Cre | ||
| + | 5. BBa_I716212: Cre(GTG Start) | ||
| + | 6. BBa_S03975: Lacl+pL Luxr+ LuxpR(Don't have plasmid) | ||
| + | 7. BBa_K200002: ygiV | ||
| + | 8. BBa_K200003: Ligase(Rfal) | ||
| + | 9. BBa_S09373: LacZ | ||
| + | 10.BBa_J31000: Invertase Hin from Salmonella Typhimurium | ||
| + | 11.BBa_B1006: Stop | ||
| + | 12.BBa_K118004: rbs+dxs | ||
| + | 13.BBa_K098995: Heat sensitive cl QPI with high promoter | ||
| - | + | Extra: | |
| - | + | BBa_J31001: DNA Invertase | |
| - | + | ||
| + | =Sequences for all the parts= | ||
| + | >BBa_K200000 Part-only sequence (654 bp) | ||
| + | atgaacaatatgaacgtaattattgccgatgaccatccgatagtcttgttcggtattcgcaaatcacttgagcaaattgagtgggtgaatgttgtcggcg | ||
| + | aatttgaagactctacagcactgatcaacaacctgccgaaactggatgcgcatgtgttgattaccgatctctccatgcctggcgataagtacggcgatgg | ||
| + | cattaccttaatcaagtacatcaagcgccatttcccaagcctgtcgatcattgttctgactatgaacaacaacccggcgattcttagtgcggtattggat | ||
| + | ctggatatcgaagggatcgtgctgaaacaaggtgcaccgaccgatctgccgaaagctctcgccgcgctccagaaagggaagaaatttaccccggaaagcg | ||
| + | tttctcgcctgttggaaaaaatcagtgctggtggttacggtgacaagcgtctctcgccaaaagagagtgaagttctgcgcctgtttgcggaaggcttcct | ||
| + | ggtgaccgagatcgctaaaaagctgaaccgcagtattaaaaccatcagtagccagaagaaatctgcgatgatgaagctgggtgtcgagaacgatatcgcc | ||
| + | ctgctgaattatctctcttcagtgaccttaagtccggcagataaagactaataa | ||
| - | |||
| - | + | >BBa_I716213 Part-only sequence (1032 bp) | |
| + | ttgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacctgatggacatgttcagggatc | ||
| + | gccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataaccggaaatggtttcccgcaga | ||
| + | acctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggccagctaaacatgcttcatcgt | ||
| + | cggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggattcgaaaagaaaacgttgatgccggtgaacgtgcaaaacagg | ||
| + | ctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaatctggcatttctggggattgc | ||
| + | ttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatgttaatccatattggcagaacg | ||
| + | aaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccgtctctggtgtagctgatgatc | ||
| + | cgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgccctggaagggatttttgaagc | ||
| + | aactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtcggagccgcgcgagatatggcc | ||
| + | cgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgtaacctggatagtgaaacagggg | ||
| + | caatggtgcgcctgctggaagatggcgattaa | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | >BBa_S04114 Part-only sequence (1410 bp) | |
| + | gattgttctatcagtaatcgaccttattcctaattaaatagagcaaatccccttattgggggtaagacatgaagatgccagaaaaacatgacctgttggc | ||
| + | cgccattctcgcggcaaaggaacaaggcatcggggcaatccttgcgtttgcaatggcgtaccttcgcggcagatataatggcggtgcgtttacaaaaaca | ||
| + | gtaatcgacgcaacgatgtgcgccattatcgcctggttcattcgtgaccttctcgacttcgccggactaagtagcaatctcgcttatataacgagcgtgt | ||
| + | ttatcggctacatcggtactgactcgattggttcgcttatcaaacgcttcgctgctaaaaaagccggagtagaagatggtagaaatcaataatcaacgta | ||
| + | aggcgttcctcgatatgctggcgtggtcggagggaactgataacggacgtcagaaaaccagaaatcatggttatgacgtcattgtaggcggagagctatt | ||
| + | tactgattactccgatcaccctcgcaaacttgtcacgctaaacccaaaactcaaatcaacaggcgccggacgctaccagcttctttcccgttggtgggat | ||
| + | gcctaccgcaagcagcttggcctgaaagacttctctccgaaaagtcaggacgctgtggcattgcagcagattaaggagcgtggcgctttacctatgattg | ||
| + | atcgtggtgatatccgtcaggcaatcgaccgttgcagcaatatctgggcttcactgccgggcgctggttatggtcagttcgagcataaggctgacagcct | ||
| + | gattgcaaaattcaaagaagcgggcggaacggtcagagagattgatgtatgagcagagtcaccgcgattatctccgctctggttatctgcatcatcgtct | ||
| + | gcctgtcatgggctgttaatcattaccgtgataacgccattacctacaaagcccagcgcgacaaaaatgccagagaactgaagctggcgaacgcggcaat | ||
| + | tactgacatgcagatgcgtcagcgtgatgttgctgcgctcgatgcaaaatacacgaaggagttagctgatgctaaagctgaaaatgatgctctgcgtgat | ||
| + | gatgttgccgctggtcgtcgtcggttgcacatcaaagcagtctgtcagtcagtgcgtgaagccaccaccgcctccggcgtggataatgcagcctcccccc | ||
| + | gactggcagacaccgctgaacgggattatttcaccctcagagagaggctgatcactatgcaaaaacaactggatactagagccaggcatcaaataaaacg | ||
| + | aaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggctcaccttcgggtgggcctttc | ||
| + | tgcgtttata | ||
| - | |||
| - | |||
| - | + | >BBa_I718007 Part-only sequence (1058 bp) | |
| + | attaaagaggagaaatactagatgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacc | ||
| + | tgatggacatgttcagggatcgccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataa | ||
| + | ccggaaatggtttcccgcagaacctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggc | ||
| + | cagctaaacatgcttcatcgtcggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggatccgaaaagaaaacgttgatg | ||
| + | ccggtgaacgtgcaaaacaggctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaa | ||
| + | tctggcatttctggggattgcttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatg | ||
| + | ttaatccatattggcagaacgaaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccg | ||
| + | tctctggtgtagctgatgatccgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgc | ||
| + | cctggaagggatttttgaagcaactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtc | ||
| + | ggagccgcgcgagatatggcccgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgta | ||
| + | acctggatagtgaaacaggggcaatggtgcgcctgctggaagatggcgattaagaatt | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | >BBa_I716212 Part-only sequence (1032 bp) | |
| - | + | ttgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacctgatggacatgttcagggatc | |
| + | gccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataaccggaaatggtttcccgcaga | ||
| + | acctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggccagctaaacatgcttcatcgt | ||
| + | cggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggattcgaaaagaaaacgttgatgccggtgaacgtgcaaaacagg | ||
| + | ctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaatctggcatttctggggattgc | ||
| + | ttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatgttaatccatattggcagaacg | ||
| + | aaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccgtctctggtgtagctgatgatc | ||
| + | cgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgccctggaagggatttttgaagc | ||
| + | aactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtcggagccgcgcgagatatggcc | ||
| + | cgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgtaacctggatagtgaaacagggg | ||
| + | caatggtgcgcctgctggaagatggcgattaa | ||
| - | |||
| - | + | >BBa_S03975 Part-only sequence (1082 bp) | |
| - | + | aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcacatactagagaaagaggagaaatactagatgaaaaacataaatgccg | |
| - | + | acgacacatacagaataattaataaaattaaagcttgtagaagcaataatgatattaatcaatgcttatctgatatgactaaaatggtacattgtgaata | |
| + | ttatttactcgcgatcatttatcctcattctatggttaaatctgatatttcaatcctagataattaccctaaaaaatggaggcaatattatgatgacgct | ||
| + | aatttaataaaatatgatcctatagtagattattctaactccaatcattcaccaattaattggaatatatttgaaaacaatgctgtaaataaaaaatctc | ||
| + | caaatgtaattaaagaagcgaaaacatcaggtcttatcactgggtttagtttccctattcatacggctaacaatggcttcggaatgcttagttttgcaca | ||
| + | ttcagaaaaagacaactatatagatagtttatttttacatgcgtgtatgaacataccattaattgttccttctctagttgataattatcgaaaaataaat | ||
| + | atagcaaataataaatcaaacaacgatttaaccaaaagagaaaaagaatgtttagcgtgggcatgcgaaggaaaaagctcttgggatatttcaaaaatat | ||
| + | taggttgcagtgagcgtactgtcactttccatttaaccaatgcgcaaatgaaactcaatacaacaaaccgctgccaaagtatttctaaagcaattttaac | ||
| + | aggagcaattgattgcccatactttaaaaattaataacactgatagtgctagtgtagatcactactagagccaggcatcaaataaaacgaaaggctcagt | ||
| + | cgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggctcaccttcgggtgggcctttctgcgtttatat | ||
| + | actagagacctgtaggatcgtacaggtttacgcaagaaaatggtttgttatagtcgaataaatactagagaaagaggagaaa | ||
| - | |||
| - | |||
| + | >BBa_K200002 Part-only sequence (486 bp) | ||
| + | atgacaaacctgacactggatgtaaacattatcgatttcccatcaatacctgtggcgatgttgccgcaccgctgtagccctgaattgctcaactacagcg | ||
| + | tggcgaaatttatcatgtggcgtaaagagacggggctttctcctgttaaccaaagccagacttttggcgtcgcctgggacgaccctgccaccaccgcacc | ||
| + | ggaagcgtttcgctttgatatctgcggcagcgttagcgaaccgattcccgataatcgttatggtgtgagcaatggtgaacttaccggtggacgttatgcc | ||
| + | gtggcccgccacgttggcgagctggacgatatttcacacacggtatggggcatcattcgccactggctgcctgcaagcggcgagaaaatgcgtaaagcac | ||
| + | cgattctgtttcactacaccaatcttgccgaaggggtgacagagcagcgactggaaacggatgtttatgtgccgttggcgtgataa | ||
| - | |||
| - | |||
| - | + | >BBa_K200003 Part-only sequence (1263 bp) | |
| - | + | atgctaacatcctttaaacttcattcattgaaaccttacactctgaaatcatcaatgattttagagataataacttatatattatgttttttttcaatga | |
| + | taattgcattcgtcgataatactttcagcataaaaatatataatatcactgctatagtttgcttattgtcactaattttacgtggcagacaagaaaatta | ||
| + | taatataaaaaaccttattcttcccctttctatatttttaataggcttgcttgatttaatttggtattctgcgtttaaagtagataattcgccatttcgt | ||
| + | gctacttaccatagttatttaaatactgccaaaatatttatatttggttcttttattgttttcttgacactaactagccagctaaaatcaaaaaaagaga | ||
| + | gtgtattatacactttgtattctctgtcatttctaattgctggatatgcaatgtatattaatagcattcatgaaaatgaccgcatttcttttggtgtagg | ||
| + | aacggcaacaggagcagcatattcaacaatgctaatagggatagttagtggcgttgcgattctttatactaagaaaaatcatccttttttatttttatta | ||
| + | aatagttgcgcggtactttatgttctggcgctaacacaaaccagagcaaccctactcctgttccctataatttgtgttgctgcattaatagcttattata | ||
| + | ataaatcacccaagaaattcacttcctctattgttctactaattgctatattagctagcattgttattatatttaataaaccaatacagaatcgctataa | ||
| + | tgaagcattaaatgacttaaacagttataccaatgctaatagtgttacttccctaggtgcaagactggcaatgtacgaaattggtttaaatatattcata | ||
| + | aagtcacctttttcatttagatcagcagagtcacgcgctgaaagtatgaatttgttagttgcagaacacaataggctaagaggggcattggagttttcta | ||
| + | acgtacatctacataatgagataattgaagcagggtcactgaaaggtctgatgggaattttttccacacttttcctctatttttcactattttatatagc | ||
| + | atataaaaaacgagctttgggtttgttgatattaacgcttggcattgtggggattggactcagtgatgtgatcatatgggcacgcagcattccaattatc | ||
| + | attatatccgctatagtcctcttactcgtcattaataatcgtaacaatacaattaattaataa | ||
| - | |||
| - | + | >BBa_S03973 Part-only sequence (3230 bp) | |
| + | aaagaggagaaatactagatgaccatgattacggattcactggccgtcgttttacaacgtcgtgactgggaaaaccctggcgttacccaacttaatcgcc | ||
| + | ttgcagcacatccccctttcgccagctggcgtaatagcgaagaggcccgcaccgatcgcccttcccaacagttgcgcagcctgaatggcgaatggcgctt | ||
| + | tgcctggtttccggcaccagaagcggtgccggaaagctggctggagtgcgatcttcctgaggccgatactgtcgtcgtcccctcaaactggcagatgcac | ||
| + | ggttacgatgcgcccatctacaccaacgtgacctatcccattacggtcaatccgccgtttgttcccacggagaatccgacgggttgttactcgctcacat | ||
| + | ttaatgttgatgaaagctggctacaggaaggccagacgcgaattatttttgatggcgttaactcggcgtttcatctgtggtgcaacgggcgctgggtcgg | ||
| + | ttacggccaggacagtcgtttgccgtctgaatttgacctgagcgcatttttacgcgccggagaaaaccgcctcgcggtgatggtgctgcgctggagtgac | ||
| + | ggcagttatctggaagatcaggatatgtggcggatgagcggcattttccgtgacgtctcgttgctgcataaaccgactacacaaatcagcgatttccatg | ||
| + | ttgccactcgctttaatgatgatttcagccgcgctgtactggaggctgaagttcagatgtgcggcgagttgcgtgactacctacgggtaacagtttcttt | ||
| + | atggcagggtgaaacgcaggtcgccagcggcaccgcgcctttcggcggtgaaattatcgatgagcgtggtggttatgccgatcgcgtcacactacgtctg | ||
| + | aacgtcgaaaacccgaaactgtggagcgccgaaatcccgaatctctatcgtgcggtggttgaactgcacaccgccgacggcacgctgattgaagcagaag | ||
| + | cctgcgatgtcggtttccgcgaggtgcggattgaaaatggtctgctgctgctgaacggcaagccgttgctgattcgaggcgttaaccgtcacgagcatca | ||
| + | tcctctgcatggtcaggtcatggatgagcagacgatggtgcaggatatcctgctgatgaagcagaacaactttaacgccgtgcgctgttcgcattatccg | ||
| + | aaccatccgctgtggtacacgctgtgcgaccgctacggcctgtatgtggtggatgaagccaatattgaaacccacggcatggtgccaatgaatcgtctga | ||
| + | ccgatgatccgcgctggctaccggcgatgagcgaacgcgtaacgcgaatggtgcagcgcgatcgtaatcacccgagtgtgatcatctggtcgctggggaa | ||
| + | tgaatcaggccacggcgctaatcacgacgcgctgtatcgctggatcaaatctgtcgatccttcccgcccggtgcagtatgaaggcggcggagccgacacc | ||
| + | acggccaccgatattatttgcccgatgtacgcgcgcgtggatgaagaccagcccttcccggctgtgccgaaatggtccatcaaaaaatggctttcgctac | ||
| + | ctggagagacgcgcccgctgatcctttgcgaatacgcccacgcgatgggtaacagtcttggcggtttcgctaaatactggcaggcgtttcgtcagtatcc | ||
| + | ccgtttacagggcggcttcgtctgggactgggtggatcagtcgctgattaaatatgatgaaaacggcaacccgtggtcggcttacggcggtgattttggc | ||
| + | gatacgccgaacgatcgccagttctgtatgaacggtctggtctttgccgaccgcacgccgcatccagcgctgacggaagcaaaacaccagcagcagtttt | ||
| + | tccagttccgtttatccgggcaaaccatcgaagtgaccagcgaatacctgttccgtcatagcgataacgagctcctgcactggatggtggcgctggatgg | ||
| + | taagccgctggcaagcggtgaagtgcctctggatgtcgctccacaaggtaaacagttgattgaactgcctgaactaccgcagccggagagcgccgggcaa | ||
| + | ctctggctcacagtacgcgtagtgcaaccgaacgcgaccgcatggtcagaagccgggcacatcagcgcctggcagcagtggcgtctggcggaaaacctca | ||
| + | gtgtgacgctccccgccgcgtcccacgccatcccgcatctgaccaccagcgaaatggatttttgcatcgagctgggtaataagcgttggcaatttaaccg | ||
| + | ccagtcaggctttctttcacagatgtggattggcgataaaaaacaactgctgacgccgctgcgcgatcagttcacccgtgcaccgctggataacgacatt | ||
| + | ggcgtaagtgaagcgacccgcattgaccctaacgcctgggtcgaacgctggaaggcggcgggccattaccaggccgaagcagcgttgttgcagtgcacgg | ||
| + | cagatacacttgctgatgcggtgctgattacgaccgctcacgcgtggcagcatcaggggaaaaccttatttatcagccggaaaacctaccggattgatgg | ||
| + | tagtggtcaaatggcgattaccgttgatgttgaagtggcgagcgatacaccgcatccggcgcggattggcctgaactgccagctggcgcaggtagcagag | ||
| + | cgggtaaactggctcggattagggccgcaagaaaactatcccgaccgccttactgccgcctgttttgaccgctgggatctgccattgtcagacatgtata | ||
| + | ccccgtacgtcttcccgagcgaaaacggtctgcgctgcgggacgcgcgaattgaattatggcccacaccagtggcgcggcgacttccagttcaacatcag | ||
| + | ccgctacagtcaacagcaactgatggaaaccagccatcgccatctgctgcacgcggaagaaggcacatggctgaatatcgacggtttccatatggggatt | ||
| + | ggtggcgacgactcctggagcccgtcagtatcggcggaatttcagctgagcgccggtcgctaccattaccagttggtctggtgtcaaaaataatactaga | ||
| + | gccaggcatcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggct | ||
| + | caccttcgggtgggcctttctgcgtttata | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | >BBa_J31000 Part-only sequence (573 bp) | |
| - | + | atggctactattgggtatattcgggtgtcaacaattgaccaaaatatcgatttacagcgtaatgcgcttaccagtgcaaattgtgaccgcatttttgaag | |
| - | + | accgtatcagtggcaagattgcaaaccgccccggcctgaaacgggcgttaaagtatgtaaataaaggcgatactcttgtcgtctggaaattagacagact | |
| - | + | gggccgtagcgtgaaaaatctggtggcgttaatatcagaattacatgaacgtggagctcacttccattctttaaccgatagtattgataccagtagcgcg | |
| - | + | atggggcgattcttttttcatgtaatgtcagcactggccgagatggagcgagaattaatcgtcgagcgaacccttgccggactggctgccgccagagcgc | |
| - | + | aaggacgactgggagggcgccctcgggcgatcaacaaacatgaacaggaacagattagtcggctattagagaaaggccatcctcggcagcaattagctat | |
| + | tatttttggtattggcgtatccaccttatacagatactttccggcaagcagtataaaaaaacgaatgaattaa | ||
| - | |||
| - | |||
| - | + | >BBa_K118004 Part-only sequence (1882 bp) | |
| + | ctcaaggaggtactagatgagttttgatattgccaaatacccgaccctggcactggtcgactccacccaggagttacgactgttgccgaaagagagttta | ||
| + | ccgaaactctgcgacgaactgcgccgctatttactcgacagcgtgagccgttccagcgggcacttcgcctccgggctgggcacggtcgaactgaccgtgg | ||
| + | cgctgcactatgtctacaacaccccgtttgaccaattgatttgggatgtggggcatcaggcttatccgcataaaattttgaccggacgccgcgacaaaat | ||
| + | cggcaccatccgtcagaaaggcggtctgcacccgttcccgtggcgcggcgaaagcgaatatgacgtattaagcgtcgggcattcatcaacctccatcagt | ||
| + | gccggaattggtattgcggttgctgccgaaaaagaaggcaaaaatcgccgcaccgtctgtgtcattggcgatggcgcgattaccgcaggcatggcgtttg | ||
| + | aagcgatgaatcacgcgggcgatatccgtcctgatatgctggtgattctcaacgacaatgaaatgtcgatttccgaaaatgtcggcgcgctcaacaacca | ||
| + | tctggcacagctgctttccggtaagctttactcttcactgcgcgaaggcgggaaaaaagttttctctggcgtgccgccaattaaagagctgctcaaacgc | ||
| + | accgaagaacatattaaaggcatggtagtgcctggcacgttgtttgaagagctgggctttaactacatcggcccggtggacggtcacgatgtgctggggc | ||
| + | ttatcaccacgctaaagaacatgcgcgacctgaaaggcccgcagttcctgcatatcatgaccaaaaaaggtcgtggttatgaaccggcagaaaaagaccc | ||
| + | gatcactttccacgccgtgcctaaatttgatccctccagcggttgtttgccgaaaagtagcggcggtttgccgagctattcaaaaatctttggcgactgg | ||
| + | ttgtgcgaaacggcagcgaaagacaacaagctgatggcgattactccggcgatgcgtgaaggttccggcatggtcgagttttcacgtaaattcccggatc | ||
| + | gctacttcgacgtggcaattgccgagcaacacgcggtgacctttgctgcgggtctggcgattggtgggtacaaacccattgtcgcgatttactccacttt | ||
| + | cctgcaacgcgcctatgatcaggtgctgcatgacgtggcgattcaaaagcttccggtcctgttcgccatcgaccgcgcgggcattgttggtgctgacggt | ||
| + | caaacccatcagggtgcttttgatctctcttacctgcgctgcataccggaaatggtcattatgaccccgagcgatgaaaacgaatgtcgccagatgctct | ||
| + | ataccggctatcactataacgatggcccgtcagcggtgcgctacccgcgtggcaacgcggtcggcgtggaactgacgccgctggaaaaactaccaattgg | ||
| + | caaaggcattgtgaagcgtcgtggcgagaaactggcgatccttaactttggtacgctgatgccagaagcggcgaaagtcgccgaatcgctgaacgccacg | ||
| + | ctggtcgatatgcgttttgtgaaaccgcttgatgaagcgttaattctggaaatggccgccagccatgaagcgctggtcaccgtagaagaaaacgccatta | ||
| + | tgggcggcgcaggcagcggcgtgaacgaagtgctgatggcccatcgtaaaccagtacccgtgctgaacattggcctgccggacttctttattccgcaagg | ||
| + | aactcaggaagaaatgcgcgccgaactcggcctcgatgccgctggtatggaagccaaaatcaaggcctggctggcataataa | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | >BBa_K098995 Part-only sequence (935 bp) | |
| + | tttatggctagctcagtcctaggtacaatgctagctactagagaaagaggagaaatactagatgagcacaaaaaagaaaccattaacacaagagcagctt | ||
| + | gaggacgcacgtcgccttaaagcaatttatgaaaaaaagaaaaatgaacttggcttatcccaggaatctgtcgcagacaagatggggatggggcagtcag | ||
| + | gcgttggtgctttatttaatggcatcaatgcattaaatgcttataacgccgcattgcttgcaaaaattctcaaagttagcgttgaagaatttagcccttc | ||
| + | aatcgccagagaaatctacgagatgtatgaagcggttagtatgcagccgtcacttagaagtgagtatgagtaccctgttttttctcatgttcaggcaggg | ||
| + | atgttctcacctgagcttagaacctttaccaaaggtgatgcggagagatgggtaagcacaaccaaaaaagccagtgattctgcattctggcttgaggttg | ||
| + | aaggtaattccatgaccgcaccaacaggctccaagccaagctttcctgacggaatgttaattctcgttgaccctgagcaggctgttgagccaggtgattt | ||
| + | ctgcatagccagacttgggggtgatgagtttaccttcaagaaactgatcagggatagcggtcaggtgtttttacaaccactaaacccacagtacccaatg | ||
| + | atcccatgcaatgagagttgttccgttgtggggaaagttatcgctagtcagtggcctgaagagacgtttggctgatactagagtcacactggctcacctt | ||
| + | cgggtgggcctttctgcgtttatatactagagagagaatataaaaagccagattattaatccggcttttttattattttactagagtaacaccgtgcgtg | ||
| + | ttgactattttacctctggcggtgataatggttgc | ||
| - | |||
| - | + | =Summary for Week June 7th to June 11th= | |
| + | 6-7-10: | ||
| + | a) Miniprep 2 pellet each for pBAD33, pBAD35, pBAD18, ygiV (attach to the cell wall), RcsB(expression factor that increase the clonic acid). | ||
| - | + | b) enzyme digestion for pBAD33, vgiV, pBAD18, and RcsB. | |
| - | + | c) Gel extract for vgiV and RcsB and purification | |
| - | + | d) PCR purification for pBAD18 and pBAD33 | |
| - | + | ||
| - | + | e) overnight ligation (set up by Sarah) | |
| - | : | + | 6-8-10: |
| - | + | a) transformation pBAD18-RcsB and pBAD33-vgiV into DH10B E. Coli. | |
| - | + | (Sarah and Hannah plates the cloning of pBAD18-RcsB and pBAD33-vgiV) | |
| - | + | ||
| - | + | b) Miniprep | |
| + | For Encryption Project: BBa_I11020, BBa_K199021, BBa_K2902006, BBa_200021, BBa_B1006-A, BBa_B1006-B, BBa_E0240 | ||
| + | For Drug Project: 2 pellets of each, pBAD18, pBAD33, pBAD35, vgiV, RcsB | ||
| - | + | 6-9-10: | |
| - | + | ||
| - | + | a) For the RcsB and VgiV plates, only a few colonies growed (they are really tiny, kindda hard to see, so we put it back to the incubator growing for couple of more hours) | |
| - | + | b) Did the cloning of RcsB into pBAD18 and vgiV into pBAD33 again with different insert/vector ratio. | |
| - | + | c) Plate the pellet and control. | |
| - | + | 6-10-10: | |
| - | + | a) The pBAD18-RcsB plate has colonies growed. | |
| - | + | ||
| - | + | b) Set up the screening with colony PCR | |
| - | + | ||
| - | + | c) Made MRS Broth, MRS plates, no antibiotic plates, Amp+Kan plates, and CM plates with Mary. | |
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 22:03, 24 October 2010
Note book of July 12th,2010
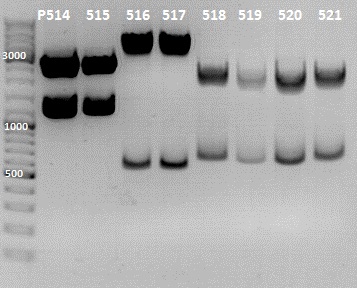
1. Screening for the Rfal into LacI-RBS cloning
Gel Picture
File:IGEM 2010-07-12 13hr 39min.jpg
File:IGEM 2010-07-12 17hr 33min.jpg
2. Primer Design For the Cloning LacI-RBS-ygiV-RBS-Rfal-RBS-RFP-Terminator
One is for Rfal: the forward primer has a XbaI and RBS in it; the reverse primer is on the backbone, and has a XmaI site on it.
pRBS-Rfal_Fwd: TGCTCTAGA AAAGAGGAGAAAATGCTAACATCCTTTAAACT
pRBS-Rfal_Rev: CCCCCCGGGTTAATTAATTGTATTGTTACGATTAT
The other one is For RFP (I13507): The forward primer has a XmaI site on it and starts from the backbone; the reverse primer starts on the backbone too. [The original plasmid, I13507 has a structur of RBS-RFP-terminator]
pRFP(I13507)_Fwd: CCCCCCGGGgagaaagaggagaaatactagatgg
pRFP(I13507)_Rev: AGTCAGTGAGCGAGGAAG
Note book of July 13th,2010
1. Primer Designs:
Tyler helped me check the primers, and he get back to me with his comments:
pRBS-RfaI_Fwd - Tm for primer portion is too low(oligocalc says 44), add 4 bases from the gene sequence, try to have 3' end be G or C
pRBS-RfaI_Rev - GC content of the priming portion is way too low, try to get it to at least above 30%, you can move down the gene since you are at the ending site, also try to have the 3' end of the primer be a G or C
pRFP(I13507)_Fwd - good, I usually add a 5 base overhang, but 3 should be ok for the enzyme you are using
pRFP(I13507)_Fwd - good
So I redesigned my primers with
Detailed information are in: File:Primer Design for Rfal and RFP - 2.pdf
Primers For Rfal
Tm : 46 GC:41% Fwd: gagatgctaacatcctt (17bp)
pRBS-RfaI_Fwd : TGCTCTAGAAAAGAGGAGAAAtacgagatgctaacatcctt
Name Sequence Tm°C CG% nt A T C seq_1 tgctctagaaaagaggagaaatacgagatgctaacatcctt 75.1 39.0 41 16.0 9.0 7.0 9.0 417600.0 12674.4 2.4 30.4
Name Sequence Tm°C CG% nt A T C G seq_1 gagatgctaacatcctt 49.9 41.2 17 5.0 5.0 4.0 3.0 164400.0 5169.4 6.1 31.4
Tm:49 GC:40% Rev: taataatactagtagcggcc (17bp)
pRBS-RfaI_Rev: GCCGCTACTAGTATTATTA [SpeI]
Name Sequence Tm°C CG% nt A T C G seq_1 gccgctactagtattatta 49.6 36.8 19 5.0 7.0 4.0 3.0 185500.0 5777.8 5.4 31.1
Primers For RFP
pRFP(I13507)_Fwd: CTAGTCTAGAgagaaagaggagaaatactagatgg [XbaI]
Name Sequence Tm°C CG% nt A T C G seq_1 ctagtctagagagaaagaggagaaatactagatgg 65.7 40.0 35 15.0 6.0 3.0 11.0 377400.0 10950.2 2.6 29.0
Name Sequence Tm°C CG% nt A T C G seq_1 gagaaagaggagaaatactagatgg 59.8 40.0 25 12.0 3.0 1.0 9.0 278000.0 7861.2 3.6 28.3
pRFP(I13507): AGTCAGTGAGCGAGGAAG (cttcctcgctcactgact)
Name Sequence Tm°C CG% nt A T C G seq_1 agtcagtgagcgaggaag 58.8 55.6 18 6.0 2.0 2.0 8.0 191900.0 5637.7 5.2 29.4
Name Sequence Tm°C CG% nt A T C G seq_1 agtcagtgagcgaggaag 58.8 55.6 18 6.0 2.0 2.0 8.0 191900.0 5637.7 5.2 29.4
Note book of July 14th,2010
Cloning of LacI-RBS-ygiV with a terminator
a. Double digestion
b. Gel extraction and purificaiton
c. overnight ligation
Note book of July 15th,2010
Cloning of LacI-RBS-Rfal with a terminator
a. Double digestion
b. Gel extraction and purificaiton
c. 2 hours bench top ligation
d. Electroporation of LacI-RBS-ygiV-terminator and LacI-RBS-Rfal-terminator into DH10B cells.
e. Plate on Amp plates
f. run a gel to check the digestion of the backbone
Note book of July 16th,2010
A.Check the plate
1. LacI-RBS-ygiV-terminator
the pallet plate has about 200 colonies
the control has 8 colonies
2.LacI-RBS-Rfal-terminator
the pallet plate has about 150 colonies
the control doesn't have colonies
B.Screening
Note book of July 6th,2010
A. Start the clone of Rfal into K200021(LacI+RBS)
1.cut the backbone, K200021, with PstI-HF and SpeI for 2 hours. cut the insert, Rfal, with PstI-HF and XbaI for 2 hours.
2.Gel Extract
K200021: 3264bp Rfal:1263bp
File:IGEM 2010-07-06 13hr 05min.jpg
After the gel extraction, the concentrations are:
K200021: 33.1ng/ul Rfal: 53.9ng/ul
3.Ligation
for doing a K200021:Rfal is 1:4 ligation, 100ng of K200021 and 154.8ng of Rfal are needed. Based on the DNA concentration, add 3ul K200021 and 3ul Rfal for ligation.
Stored at 16C overnight.
Note book of July 7th,2010
A. Cloning of LacI-RBS-Rfal
1.Electroporation
LacI-RBS-Rfal has a time constant 4.8;
The control has a time constant 4.7;
2.Plate on two Amp+Kan plate. Stored in a 37C incubator.
B. Cloning of LacI-RBS-ygiV into RBS
1.cut the backbone, RBS, with EcoRI-HF and XbaI, and cut the insert LacI-RBS-ygiV with EcoRI-HF and SpeI.
2.Gel Extraction
RBS:3200bp LacI-RBS-ygiV:543bp
File:IGEM 2010-07-06 17hr 00min.jpg
after the gel extraction, the concentrations are:
RBS: 20.7ng/ul
lacI-RBS-ygiV: 19.8ng/ul
3.Ligation
For a 1 to 3 backbone:insert ratio, we need 100ng of RBS and 107.3ng of LacI-RBS-ygiV for Ligation. THerefore, add 3.7ul RBS and 4.3ul LacI-RBS-ygiV to ligate. Store at 4C overnight.
C.Run a gel to check the cutting peices
The orders are:
L-ygiV-ES; L-ygiV uncut; Rfal-PX; Rfal uncut; RBS-EX; RBS uncut; LacI-PS; LacI uncut; Ladder
unluckily, the first lane for L-ygiV-ES didn't show up any band.
File:IGEM 2010-07-07 11hr 24min.jpg
The rest of the bands are all correct.
Then run a gel for L-ygiV-ES and L-ygiV uncut by loading more DNA on the gel
File:IGEM 2010-07-07 13hr 09min.jpg
Now, this shows that the L-ygiV-ES is correct.
Note book of July 8th,2010
A. Transformation of the Clone LacI-RBS-ygiV-RBS
1. Electroporation
LacI-RBS-ygiV-RBS: 4.7
Control: 4.9
2. Plate on two Amp plate.
B. Run a gel to check the Ligation of LacI-RBS-Rfal and LacI-RBS-ygiV-RBS
the orders are Ladder LacI-RBS-Rfal, control, LacI-RBS-ygiV-RBS, control
File:IGEM 2010-07-07 12hr 48min.jpg
C. Colony PCR for the LacI-RBS-Rfal Clone
1.The control plate only has one colony, compared with the regular plate with about 100 colonies.
2.picked 14 colonies for screening.
Gel for Colony PCR:
File:IGEM 2010-07-08 13hr 54min.jpg
Run for 5mins longer and add more EB
File:IGEM 2010-07-08 14hr 20min.jpg
No colony shows the correct band (around 1500bp)
Note book of July 9th,2010
A Cloning of LacI-RBS-ygiV into RBS, and Rfal into LacI-RBS.
Add 1.5mg of DNA into each tube:
RBS(B0034):5.3ul template; Add 1ul EcoRI-HF to digest 1 hour and inactivate at 65C for 20mins. Then Add 1ul of XbaI to digest an additional hour.
LacI-RBS(K200021): 9.5ul template; Add 1ul PstI-HF to digest 1 hour and inactivate at 80C for 20mins. Then Add 1ul of SpeI to digest an additional hour.
Rfal: 3.1ul template digest with 1ul PstI-HF and 1ul XbaI
LacI-ygiV: 4.4ul tempalte digest with 1ul EcoRI-HF and 1ul SpeI
Gel after 1 hour inactivation of EcoRI-HF in RBS and PstI-HF in LacI-RBS:
Orders are:
RBS cut with EcoRI-HF, RBS uncut, Ladder, LacI-RBS cut with PstI-HF, LacI-RBS uncut.
File:IGEM 2010-07-09 13hr 27min.jpg
Note book of June 28th,2010
a. Digestion of K200021 (LacI+RBS) with PstI-HF and SpeI, and digest ygiV and Rfal with PstI-HF and XbaI.
b. Gel Extraction and purification:
1. Gel
K200021-PS
ygiV and Rfal
File:IGEM 2010-06-27 13hr 19min.jpg
Concentration of the cutting peices: K200021-PS
ygiV-PX
Rfal-PX
Gel for the purification product
File:IGEM 2010-06-28 14hr 10min.jpg
c. Set up an overnight ligation
Note book of June 29th,2010
a. Transformation
pLacI-RBS-ygiV-PX: 4.1
pLacI-RBS-ygiV-PX(C): 3.9
pLacI-RBS-Rfal-PX: 4.0
pLacI-RBS-Rfal-PX(C):3.9
b. Plate on Amp100 plates.
Note book of June 30th,2010
Re-plan the project. Figure out all the cloning need to be done by the end of the summer. Detailed information please see the spread sheet on Sarah's page.
Note book of July 1st,2010
A. Colony PCR of the LacI-RBS-ygiV and LacI-RBS-Rfal clone.
Upper: LacI-RBS-Rfal Clone
Lower: LacI-RBS-ygiV clone
File:IGEM 2010-07-01 14hr 49min-2.jpg
The LacI-RBS-ygiV clone shows up at the right length.
B. Innoculate 5 liquid culture of the ygiV clone.
Note book of July 2nd,2010
A. Made 3 freezer stocks of LacI-RBS-ygiV clone
B. Miniprep LacI-RBS-ygiV
LacI-RBS-ygiV-1: 388.7ng/ul LacI-RBS-ygiV-2: 357.0ng/ul LacI-RBS-ygiV-3: 350.2ng/ul LacI-RBS-ygiV-4: 255.0ng/ul
Note book of June 21st,2010
a.Minipreped 3 sets each for pBAD33BB, pBAD35BB, ygiV, and Rfal.
b.Set up a overnight digestions that cut pBAD33BB and pBAD35BB with PstI-HF and SpeI, and cut ygiV and Rfal with PstI-HF and XbaI. (37C overnight)
c.Screening for the 2nd time ygiV and Rfal cloning.
ygiV:
File:IGEM 2010-06-21 bads2 Peter ScanA.jpg
Rfal:
File:IGEM 2010-06-21 bads2 Peter ScanB.jpg
They all shows up at the same length as the positive control, which means we didn't get the desired cloning.
Note book of June 22nd,2010
a.Gel extraction of pBAD33BB, ygiV and Rfal.
For pBAD33BB
File:IGEM 2010-06-23 12hr 45min.jpg
For ygiV(486bp) and Rfal(1269bp)
File:IGEM 2010-06-23 12hr 44min.jpg
b.Gel purification of pBAD33BB, ygiV and Rfal.
after the gel purificaion, the concentrations are:
pBAD33BB-PX: 39.7ng/ul
ygiV: 25.7ng/ul
Rfal: 33.5ng/ul
c.Set up an overnight ligation with Insert/Vector ratio equals 4.
tubeA
pBAD33BB: 3.8ul
ygiV: 2.2ul
tubeB
pBAD33BB: 3.8ul
Rfal: 4.2ul
tube C (control)
pBAD33BB: 3.8ul
(16C overnight)
Note book of June 23rd,2010
a. Transformation
pBAD33BB-ygiV: 4.4 1.8
pBAD33BB-Rfal: 4.4 1.8
Control: 4.3 1.8
b. Plating on CM plate
c. run the gel for ligation product
File:Pflegerlab 2010-06-24 11hr 07min.jpg
Note book of June 24th,2010
a. Check the transformation plate
pBAD33BB-ygiV:
pBAD33BB-Rfal:
Control:
b. pick 12 colonies from pBAD33BB-Rfal plate and 10 colonies from pBAD33BB-ygiV plate for the colony PCR screening.
ygiV-plate B
File:Plate B of ygiV-pBAD33BB screening pflegerlab 2010-06-25 14hr 45min.jpg
Rfal-plate B
File:Plate A of Rfal-pBAD33BB screening pflegerlab 2010-06-25 14hr 42min.jpg
c. pick colonies from both plates into liquid cultures. Grow overnight. (5 from plateA and 5 from plateB)
Note book of June 25th,2010
a. miniprep 10 liquid culture for Screening.
b. Digest with BamHI-HF and PstI-HF
c. run the gel for the screening
Summary of Week June 14th to June 18th, 2010
06-14-10
A. Miniprep
Miniprep All the new parts received
1. K082006 52.7ng/ul
2. K142001 196.7ng/ul
3. K173004 330.2ng/ul
4. pBAD35BB 102.4ng/ul
5. pBAD33BB 61.5ng/ul
6. pBAD35BB 105.8ng/ul
7. K142003 212.8ng/ul
8. K137113 174.8ng/ul
9. K142002 222.5ng/ul
10.pBAD35BB 117.2ng/ul
11.K200003 207.2ng/ul
12.K142000 177.4ng/ul
B. Ligation for vgiV and pBAD33BB
Ligation of vgiV and pBAD33BB with a molar ratio 3:1
one for 2 hours, one for overnight
C. Thansformation of the vgiV cloning
Transformed the 2-hour ligation and plated
06-15-10
A. Screening of the cloning of vgiV into pBAD33BB
The 2-hour ligation only had two colonies. Screening with Colony PCR but didn't see any band.
B. Transformation of vgiV cloning
Transformed the overnight ligation and plated in CM argar plate.
C. Restriction Mapping of the Newly Receiving Parts
The Correct Parts are
Parts Band 1@ Band2@ Band3@
K112808 2500 1406
K137113 2200 650
K173004 3200 2100 1106
K142000 2100 1100
K142001 2100 1100
K142002 2100 1100
K142003 2100 1100
K082006 3200 750
K200003 3200 1300
06-16-10
A. Screening of the cloning of vgiV into pBAD33BB
Saw a lot colony on both the 100ul plate and the pallet plate. The Control plate (backbone only) only had one colony.
Pick up 10 colonies from each plate( 100ul plate and pallet plate). Colony PCR for the screening
Run the Gel for the Coloy PCR, but didn't see the correct band.
(The band is about 200bp, which means the back bone ligate itself.)
B. Cloning of Rfal into pBAD35BB
Digest pBAD35BB with PstI-HF and XbaI, and digest Rfal with PstI-HF and SpeI
Gel extraction and purification.
Ligate with a insert/vector ratio 3:1
Ligate at 16C overnight
06-17-10
A. Another Screening of the cloning of vgiV into pBAD33BB
Pick another 10 colonies from both the 100ul plate and the pallet plate.
Proceed the Colony PCR
Run the Gel, but still didn't see the correct band(about 700bp).
Do see a band down at 200bp, same length as the positive control. Therefore, confirmed that the colonies are just transformed with the backbone.
B. Cloning of Rfal into pBAD35BB
Transform the Rfal cloning and the control into DH10B
Plated on Kan plate.
Cloning of pET28b+RFP
File:Cloning of pET28b-RFP.pdf
Notebook May 25th, 2010
BioBrick pBAD35 a) Target Vector: pBAD35 (about 4200bp, 4.2kb)
pBAD BioBrick Primers:
Tm value: Fwd: 72.0C
Rev: 68.1C
b) General Procedure 1. PCR amplify 2. Pour 2 gels during this time 3. Check PCR on 1st Gel 4. Add 1ul DpnI/rxn for 1hr at 37C 5. PCR clean up or gel extraction (depends on whether you see a bright band at right place) 6. XbarI Digest 7. PCR clean up 8. Ligate
c) Cycle Design 98C 1min 98C 15s 73C 30s 33X 72c 126S 72c 5mins 4c forever
d)Phusion Mix Water: 35ul 5Xbuffer:10ul template:2ul Primer FW:0.5ul Primer RV:0.5ul dNTP:1ul Phusion Enzyme:1ul
Note: Keep the primers and Enzyme on ICE
RESULT 1st Gel Lane Band@ 35A 3-4kb 35B 3-4kb 18 4kb 33 5kb 34 4kb and 2.5kb(wrong)
File:IGEM Biobrick first 2010-05-25 15hr 09min.jpg
Protocol of PCR Purification: File:PCR purification-Qiagen.pdf
DIGESTION
pBAD35-A DNA: 40ul Buffer4: 5ul BSA: 0.5ul Water: 3.5ul XbaI: 1ul
pBAD35-B DNA: 40ul Buffer4: 5ul BSA: 0.5ul Water: 2.5ul XbaI: 2ul
Leave the digestion on a 37c water bath overnight
Inoculation for Encryption Project
10ml LB in each tube and add 10ul Antibiotic A: BBa_K118004 B: BBa_K098995 C: BBa_I718008 D: BBa_B1006
Notebook May 26th, 2010
a)PCR purification(refer to protocol on the protocol from Notebook May 25th 2010) DNA Conc(ng/ul) pBAD35-A 57.6 pBAD35-B 69.7
b)Ligation (Peter did all the ligation) Ligate on pBAD18,33,34,35A-B 100ng/10ul rxn 2hrs bench ligation
c)Electroporation transformation
Use DH10B Add 1ul of plasmid Let it sits on ice for 5 mins
after the shock, immediately add 950ul LB and mix well Transfer to centrifuge tube
DNA Time 35 4.2 33 4.4 18 4.2
(control) 35 4.5 33 4.2 18 4.2
Let them grow in the 37c shaker for 1hr.
Plate 50ul of cells
Miniprep for Nate Pant's Project Miniprep Plasmid information A: BBa_K118004 B: BBa_K098995 C: BBa_I718008 D: BBa_B1006
Centrifuge at 30000rpm for 10mins
Follow the Miniprep Protocol: File:Qiagenmini.pdf Result Measured by Nanodrop
DNA Conc(ng/ul) 260/280 A 324.0 1.97 B 288.3 1.98 C 418.2 1.96 D 276.6 1.97
Notebook May 27th, 2010
Check the colonies in the plates.
Picked 10 colonies from pBAD35B Picked 5 colonies from pBAD33 Picked 5 colonies from pBAD18
Inoculate them in 5ml LB with antibiotics
Parts Checking
Concentration: BBa_K200000: 199.9ng/ul BBa_I716213: 186.5ng/ul BBa_S04114: 543.6ng/ul BBa_I718007: 146.1ng/ul BBa_I716212: 155.8ng/ul BBa_S03975: BBa_K200002: 309.3ng/ul BBa_K200003: 105.4ng/ul BBa_S09373: 243.1ng/ul BBa_J31000: 278.3ng/ul BBa_B1006: 276.6ng/ul BBa_K118004: 324.0ng/ul BBa_K098995: 288.3ng/ul
Single Digestion Test With EcoRI for 1. BBa_K200000: 2. BBa_I716213: 3. BBa_S04114: 4. BBa_I718007: 5. BBa_I716212: 6. BBa_S03975:(Don't have plasmid) 7. BBa_K200002: 8. BBa_K200003: 9. BBa_S09373: 10.BBa_J31000: 11.BBa_B1006: 12.BBa_K118004: 13.BBa_K098995:
Protocol 1.8ul Buffer3(10X) 0.18ul BSA(100X) 12ul MiliQ H2O 3ul DNA 1ul EcoRI
Incubate in water bath at 37C for 2 hours (total volume 18ul) add 3.6ul Dye (6X)
Load 6ul into the GEL Gel: File:2010-05-27 Map - Single digest.jpg
Gel Description: 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 12, 13
Results: 3, 9, 11, and 12 don't have the correct bands
So run the single digestion for all four again with 2 hours Gel: File:IGEM Single Digestion Second 2010-05-28 14hr 39min.jpg
Gel Description: 3, 9, Ladder, 11, 12
Result: Still not correct
Double Digestion Find one enzyme cutting site with the gene 1. BBa_K200000: Buffer:4 Enzyme:AflII EcoRI 2. BBa_I716213: Buffer:2 Enzyme:AflIII SpeI 3. BBa_S04114: Buffer:3 Enzyme:EcoRV EcoRI 4. BBa_I718007: Buffer:3 Enzyme:EcoRV EcoRI 5. BBa_I716212: Buffer:3 Enzyme:EcoRV EcoRI 6. BBa_S03975:(Don't have plasmid) 7. BBa_K200002: (don't have enzyme stock that can cut within the gene) 8. BBa_K200003: Buffer:4 Enzyme:NheI EcoRI 9. BBa_S09373: Buffer:3 Enzyme:EcoRV EcoRI 10.BBa_J31000: Buffer:3 Enzyme:BglI EcoRI 11.BBa_B1006: (Too small, cannot cut within the gene) 12.BBa_K118004: Buffer:3 Enzyme:EcoRV EcoRI 13.BBa_K098995: Buffer:2 Enzyme:HindIII EcoRI
Protocol 1.8ul NEB Buffer(10X) 0.18ul BSA(100X) 11ul MiliQ H2O 3ul DNA 1ul Enzyme1 1ul Enzyme2
Incubate in water bath at 37C for 2 hours (total volume 18ul) add 3.6ul Dye (6X)
Load 6ul into the GEL
Gel File:IGEM Parts Double Digestion 2010-05-28 14hr 54min.jpg File:IGEM Parts Double Digestion White 2010-05-28 14hr 57min.jpg
Result Only 1, 2, 4, 5, 8, 13 show the correct bands
Notebook May 28th, 2010
Miniprep for the screening
Restriction Enzyme Mapping for all the 20 screening plasmid
1.8ul Buffer3(10X) 0.18ul BSA(100X) 12ul MiliQ H2O 3ul DNA 1ul SpeI
Incubate in water bath at 37C for 2 hours (total volume 18ul) add 3.6ul Dye (6X)
Load 6ul into the GEL
Run the GEL at 100V for 30mins File:IGEM Biobrick 2010-05-28 16hr 55min.jpg GEL upper 18-1 18-2 18-3 18-4 18-5 Ladder 33-1 33-2 33-3 33-4 33-5
Gel Lower 35-1 35-2 35-3 35-4 35-5 Ladder 35-6 35-7 35-8 35-9 35-10
Notebook June 1st, 2010
Search the Detailed info of the Parts we received 1. BBa_K200000: Colanic acid global regulator (RcsB) 2. BBa_I716213: Cre(TTG Start) 3. BBa_S04114: Lysis 4. BBa_I718007: RBS-Cre 5. BBa_I716212: Cre(GTG Start) 6. BBa_S03975: Lacl+pL Luxr+ LuxpR(Don't have plasmid) 7. BBa_K200002: ygiV 8. BBa_K200003: Ligase(Rfal) 9. BBa_S09373: LacZ 10.BBa_J31000: Invertase Hin from Salmonella Typhimurium 11.BBa_B1006: Stop 12.BBa_K118004: rbs+dxs 13.BBa_K098995: Heat sensitive cl QPI with high promoter
Extra: BBa_J31001: DNA Invertase
Sequences for all the parts
>BBa_K200000 Part-only sequence (654 bp) atgaacaatatgaacgtaattattgccgatgaccatccgatagtcttgttcggtattcgcaaatcacttgagcaaattgagtgggtgaatgttgtcggcg aatttgaagactctacagcactgatcaacaacctgccgaaactggatgcgcatgtgttgattaccgatctctccatgcctggcgataagtacggcgatgg cattaccttaatcaagtacatcaagcgccatttcccaagcctgtcgatcattgttctgactatgaacaacaacccggcgattcttagtgcggtattggat ctggatatcgaagggatcgtgctgaaacaaggtgcaccgaccgatctgccgaaagctctcgccgcgctccagaaagggaagaaatttaccccggaaagcg tttctcgcctgttggaaaaaatcagtgctggtggttacggtgacaagcgtctctcgccaaaagagagtgaagttctgcgcctgtttgcggaaggcttcct ggtgaccgagatcgctaaaaagctgaaccgcagtattaaaaccatcagtagccagaagaaatctgcgatgatgaagctgggtgtcgagaacgatatcgcc ctgctgaattatctctcttcagtgaccttaagtccggcagataaagactaataa
>BBa_I716213 Part-only sequence (1032 bp)
ttgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacctgatggacatgttcagggatc
gccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataaccggaaatggtttcccgcaga
acctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggccagctaaacatgcttcatcgt
cggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggattcgaaaagaaaacgttgatgccggtgaacgtgcaaaacagg
ctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaatctggcatttctggggattgc
ttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatgttaatccatattggcagaacg
aaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccgtctctggtgtagctgatgatc
cgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgccctggaagggatttttgaagc
aactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtcggagccgcgcgagatatggcc
cgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgtaacctggatagtgaaacagggg
caatggtgcgcctgctggaagatggcgattaa
>BBa_S04114 Part-only sequence (1410 bp) gattgttctatcagtaatcgaccttattcctaattaaatagagcaaatccccttattgggggtaagacatgaagatgccagaaaaacatgacctgttggc cgccattctcgcggcaaaggaacaaggcatcggggcaatccttgcgtttgcaatggcgtaccttcgcggcagatataatggcggtgcgtttacaaaaaca gtaatcgacgcaacgatgtgcgccattatcgcctggttcattcgtgaccttctcgacttcgccggactaagtagcaatctcgcttatataacgagcgtgt ttatcggctacatcggtactgactcgattggttcgcttatcaaacgcttcgctgctaaaaaagccggagtagaagatggtagaaatcaataatcaacgta aggcgttcctcgatatgctggcgtggtcggagggaactgataacggacgtcagaaaaccagaaatcatggttatgacgtcattgtaggcggagagctatt tactgattactccgatcaccctcgcaaacttgtcacgctaaacccaaaactcaaatcaacaggcgccggacgctaccagcttctttcccgttggtgggat gcctaccgcaagcagcttggcctgaaagacttctctccgaaaagtcaggacgctgtggcattgcagcagattaaggagcgtggcgctttacctatgattg atcgtggtgatatccgtcaggcaatcgaccgttgcagcaatatctgggcttcactgccgggcgctggttatggtcagttcgagcataaggctgacagcct gattgcaaaattcaaagaagcgggcggaacggtcagagagattgatgtatgagcagagtcaccgcgattatctccgctctggttatctgcatcatcgtct gcctgtcatgggctgttaatcattaccgtgataacgccattacctacaaagcccagcgcgacaaaaatgccagagaactgaagctggcgaacgcggcaat tactgacatgcagatgcgtcagcgtgatgttgctgcgctcgatgcaaaatacacgaaggagttagctgatgctaaagctgaaaatgatgctctgcgtgat gatgttgccgctggtcgtcgtcggttgcacatcaaagcagtctgtcagtcagtgcgtgaagccaccaccgcctccggcgtggataatgcagcctcccccc gactggcagacaccgctgaacgggattatttcaccctcagagagaggctgatcactatgcaaaaacaactggatactagagccaggcatcaaataaaacg aaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggctcaccttcgggtgggcctttc tgcgtttata
>BBa_I718007 Part-only sequence (1058 bp) attaaagaggagaaatactagatgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacc tgatggacatgttcagggatcgccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataa ccggaaatggtttcccgcagaacctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggc cagctaaacatgcttcatcgtcggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggatccgaaaagaaaacgttgatg ccggtgaacgtgcaaaacaggctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaa tctggcatttctggggattgcttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatg ttaatccatattggcagaacgaaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccg tctctggtgtagctgatgatccgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgc cctggaagggatttttgaagcaactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtc ggagccgcgcgagatatggcccgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgta acctggatagtgaaacaggggcaatggtgcgcctgctggaagatggcgattaagaatt
>BBa_I716212 Part-only sequence (1032 bp) ttgtccaatttactgaccgtacaccaaaatttgcctgcattaccggtcgatgcaacgagtgatgaggttcgcaagaacctgatggacatgttcagggatc gccaggcgttttctgagcatacctggaaaatgcttctgtccgtttgccggtcgtgggcggcatggtgcaagttgaataaccggaaatggtttcccgcaga acctgaagatgttcgcgattatcttctatatcttcaggcgcgcggtctggcagtaaaaactatccagcaacatttgggccagctaaacatgcttcatcgt cggtccgggctgccacgaccaagtgacagcaatgctgtttcactggttatgcggcggattcgaaaagaaaacgttgatgccggtgaacgtgcaaaacagg ctctagcgttcgaacgcactgatttcgaccaggttcgttcactcatggaaaatagcgatcgctgccaggatatacgtaatctggcatttctggggattgc ttataacaccctgttacgtatagccgaaattgccaggatcagggttaaagatatctcacgtactgacggtgggagaatgttaatccatattggcagaacg aaaacgctggttagcaccgcaggtgtagagaaggcacttagcctgggggtaactaaactggtcgagcgatggatttccgtctctggtgtagctgatgatc cgaataactacctgttttgccgggtcagaaaaaatggtgttgccgcgccatctgccaccagccagctatcaactcgcgccctggaagggatttttgaagc aactcatcgattgatttacggcgctaaggatgactctggtcagagatacctggcctggtctggacacagtgcccgtgtcggagccgcgcgagatatggcc cgcgctggagtttcaataccggagatcatgcaagctggtggctggaccaatgtaaatattgtcatgaactatatccgtaacctggatagtgaaacagggg caatggtgcgcctgctggaagatggcgattaa
>BBa_S03975 Part-only sequence (1082 bp)
aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcacatactagagaaagaggagaaatactagatgaaaaacataaatgccg
acgacacatacagaataattaataaaattaaagcttgtagaagcaataatgatattaatcaatgcttatctgatatgactaaaatggtacattgtgaata
ttatttactcgcgatcatttatcctcattctatggttaaatctgatatttcaatcctagataattaccctaaaaaatggaggcaatattatgatgacgct
aatttaataaaatatgatcctatagtagattattctaactccaatcattcaccaattaattggaatatatttgaaaacaatgctgtaaataaaaaatctc
caaatgtaattaaagaagcgaaaacatcaggtcttatcactgggtttagtttccctattcatacggctaacaatggcttcggaatgcttagttttgcaca
ttcagaaaaagacaactatatagatagtttatttttacatgcgtgtatgaacataccattaattgttccttctctagttgataattatcgaaaaataaat
atagcaaataataaatcaaacaacgatttaaccaaaagagaaaaagaatgtttagcgtgggcatgcgaaggaaaaagctcttgggatatttcaaaaatat
taggttgcagtgagcgtactgtcactttccatttaaccaatgcgcaaatgaaactcaatacaacaaaccgctgccaaagtatttctaaagcaattttaac
aggagcaattgattgcccatactttaaaaattaataacactgatagtgctagtgtagatcactactagagccaggcatcaaataaaacgaaaggctcagt
cgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggctcaccttcgggtgggcctttctgcgtttatat
actagagacctgtaggatcgtacaggtttacgcaagaaaatggtttgttatagtcgaataaatactagagaaagaggagaaa
>BBa_K200002 Part-only sequence (486 bp) atgacaaacctgacactggatgtaaacattatcgatttcccatcaatacctgtggcgatgttgccgcaccgctgtagccctgaattgctcaactacagcg tggcgaaatttatcatgtggcgtaaagagacggggctttctcctgttaaccaaagccagacttttggcgtcgcctgggacgaccctgccaccaccgcacc ggaagcgtttcgctttgatatctgcggcagcgttagcgaaccgattcccgataatcgttatggtgtgagcaatggtgaacttaccggtggacgttatgcc gtggcccgccacgttggcgagctggacgatatttcacacacggtatggggcatcattcgccactggctgcctgcaagcggcgagaaaatgcgtaaagcac cgattctgtttcactacaccaatcttgccgaaggggtgacagagcagcgactggaaacggatgtttatgtgccgttggcgtgataa
>BBa_K200003 Part-only sequence (1263 bp) atgctaacatcctttaaacttcattcattgaaaccttacactctgaaatcatcaatgattttagagataataacttatatattatgttttttttcaatga taattgcattcgtcgataatactttcagcataaaaatatataatatcactgctatagtttgcttattgtcactaattttacgtggcagacaagaaaatta taatataaaaaaccttattcttcccctttctatatttttaataggcttgcttgatttaatttggtattctgcgtttaaagtagataattcgccatttcgt gctacttaccatagttatttaaatactgccaaaatatttatatttggttcttttattgttttcttgacactaactagccagctaaaatcaaaaaaagaga gtgtattatacactttgtattctctgtcatttctaattgctggatatgcaatgtatattaatagcattcatgaaaatgaccgcatttcttttggtgtagg aacggcaacaggagcagcatattcaacaatgctaatagggatagttagtggcgttgcgattctttatactaagaaaaatcatccttttttatttttatta aatagttgcgcggtactttatgttctggcgctaacacaaaccagagcaaccctactcctgttccctataatttgtgttgctgcattaatagcttattata ataaatcacccaagaaattcacttcctctattgttctactaattgctatattagctagcattgttattatatttaataaaccaatacagaatcgctataa tgaagcattaaatgacttaaacagttataccaatgctaatagtgttacttccctaggtgcaagactggcaatgtacgaaattggtttaaatatattcata aagtcacctttttcatttagatcagcagagtcacgcgctgaaagtatgaatttgttagttgcagaacacaataggctaagaggggcattggagttttcta acgtacatctacataatgagataattgaagcagggtcactgaaaggtctgatgggaattttttccacacttttcctctatttttcactattttatatagc atataaaaaacgagctttgggtttgttgatattaacgcttggcattgtggggattggactcagtgatgtgatcatatgggcacgcagcattccaattatc attatatccgctatagtcctcttactcgtcattaataatcgtaacaatacaattaattaataa
>BBa_S03973 Part-only sequence (3230 bp)
aaagaggagaaatactagatgaccatgattacggattcactggccgtcgttttacaacgtcgtgactgggaaaaccctggcgttacccaacttaatcgcc
ttgcagcacatccccctttcgccagctggcgtaatagcgaagaggcccgcaccgatcgcccttcccaacagttgcgcagcctgaatggcgaatggcgctt
tgcctggtttccggcaccagaagcggtgccggaaagctggctggagtgcgatcttcctgaggccgatactgtcgtcgtcccctcaaactggcagatgcac
ggttacgatgcgcccatctacaccaacgtgacctatcccattacggtcaatccgccgtttgttcccacggagaatccgacgggttgttactcgctcacat
ttaatgttgatgaaagctggctacaggaaggccagacgcgaattatttttgatggcgttaactcggcgtttcatctgtggtgcaacgggcgctgggtcgg
ttacggccaggacagtcgtttgccgtctgaatttgacctgagcgcatttttacgcgccggagaaaaccgcctcgcggtgatggtgctgcgctggagtgac
ggcagttatctggaagatcaggatatgtggcggatgagcggcattttccgtgacgtctcgttgctgcataaaccgactacacaaatcagcgatttccatg
ttgccactcgctttaatgatgatttcagccgcgctgtactggaggctgaagttcagatgtgcggcgagttgcgtgactacctacgggtaacagtttcttt
atggcagggtgaaacgcaggtcgccagcggcaccgcgcctttcggcggtgaaattatcgatgagcgtggtggttatgccgatcgcgtcacactacgtctg
aacgtcgaaaacccgaaactgtggagcgccgaaatcccgaatctctatcgtgcggtggttgaactgcacaccgccgacggcacgctgattgaagcagaag
cctgcgatgtcggtttccgcgaggtgcggattgaaaatggtctgctgctgctgaacggcaagccgttgctgattcgaggcgttaaccgtcacgagcatca
tcctctgcatggtcaggtcatggatgagcagacgatggtgcaggatatcctgctgatgaagcagaacaactttaacgccgtgcgctgttcgcattatccg
aaccatccgctgtggtacacgctgtgcgaccgctacggcctgtatgtggtggatgaagccaatattgaaacccacggcatggtgccaatgaatcgtctga
ccgatgatccgcgctggctaccggcgatgagcgaacgcgtaacgcgaatggtgcagcgcgatcgtaatcacccgagtgtgatcatctggtcgctggggaa
tgaatcaggccacggcgctaatcacgacgcgctgtatcgctggatcaaatctgtcgatccttcccgcccggtgcagtatgaaggcggcggagccgacacc
acggccaccgatattatttgcccgatgtacgcgcgcgtggatgaagaccagcccttcccggctgtgccgaaatggtccatcaaaaaatggctttcgctac
ctggagagacgcgcccgctgatcctttgcgaatacgcccacgcgatgggtaacagtcttggcggtttcgctaaatactggcaggcgtttcgtcagtatcc
ccgtttacagggcggcttcgtctgggactgggtggatcagtcgctgattaaatatgatgaaaacggcaacccgtggtcggcttacggcggtgattttggc
gatacgccgaacgatcgccagttctgtatgaacggtctggtctttgccgaccgcacgccgcatccagcgctgacggaagcaaaacaccagcagcagtttt
tccagttccgtttatccgggcaaaccatcgaagtgaccagcgaatacctgttccgtcatagcgataacgagctcctgcactggatggtggcgctggatgg
taagccgctggcaagcggtgaagtgcctctggatgtcgctccacaaggtaaacagttgattgaactgcctgaactaccgcagccggagagcgccgggcaa
ctctggctcacagtacgcgtagtgcaaccgaacgcgaccgcatggtcagaagccgggcacatcagcgcctggcagcagtggcgtctggcggaaaacctca
gtgtgacgctccccgccgcgtcccacgccatcccgcatctgaccaccagcgaaatggatttttgcatcgagctgggtaataagcgttggcaatttaaccg
ccagtcaggctttctttcacagatgtggattggcgataaaaaacaactgctgacgccgctgcgcgatcagttcacccgtgcaccgctggataacgacatt
ggcgtaagtgaagcgacccgcattgaccctaacgcctgggtcgaacgctggaaggcggcgggccattaccaggccgaagcagcgttgttgcagtgcacgg
cagatacacttgctgatgcggtgctgattacgaccgctcacgcgtggcagcatcaggggaaaaccttatttatcagccggaaaacctaccggattgatgg
tagtggtcaaatggcgattaccgttgatgttgaagtggcgagcgatacaccgcatccggcgcggattggcctgaactgccagctggcgcaggtagcagag
cgggtaaactggctcggattagggccgcaagaaaactatcccgaccgccttactgccgcctgttttgaccgctgggatctgccattgtcagacatgtata
ccccgtacgtcttcccgagcgaaaacggtctgcgctgcgggacgcgcgaattgaattatggcccacaccagtggcgcggcgacttccagttcaacatcag
ccgctacagtcaacagcaactgatggaaaccagccatcgccatctgctgcacgcggaagaaggcacatggctgaatatcgacggtttccatatggggatt
ggtggcgacgactcctggagcccgtcagtatcggcggaatttcagctgagcgccggtcgctaccattaccagttggtctggtgtcaaaaataatactaga
gccaggcatcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggct
caccttcgggtgggcctttctgcgtttata
>BBa_J31000 Part-only sequence (573 bp) atggctactattgggtatattcgggtgtcaacaattgaccaaaatatcgatttacagcgtaatgcgcttaccagtgcaaattgtgaccgcatttttgaag accgtatcagtggcaagattgcaaaccgccccggcctgaaacgggcgttaaagtatgtaaataaaggcgatactcttgtcgtctggaaattagacagact gggccgtagcgtgaaaaatctggtggcgttaatatcagaattacatgaacgtggagctcacttccattctttaaccgatagtattgataccagtagcgcg atggggcgattcttttttcatgtaatgtcagcactggccgagatggagcgagaattaatcgtcgagcgaacccttgccggactggctgccgccagagcgc aaggacgactgggagggcgccctcgggcgatcaacaaacatgaacaggaacagattagtcggctattagagaaaggccatcctcggcagcaattagctat tatttttggtattggcgtatccaccttatacagatactttccggcaagcagtataaaaaaacgaatgaattaa
>BBa_K118004 Part-only sequence (1882 bp) ctcaaggaggtactagatgagttttgatattgccaaatacccgaccctggcactggtcgactccacccaggagttacgactgttgccgaaagagagttta ccgaaactctgcgacgaactgcgccgctatttactcgacagcgtgagccgttccagcgggcacttcgcctccgggctgggcacggtcgaactgaccgtgg cgctgcactatgtctacaacaccccgtttgaccaattgatttgggatgtggggcatcaggcttatccgcataaaattttgaccggacgccgcgacaaaat cggcaccatccgtcagaaaggcggtctgcacccgttcccgtggcgcggcgaaagcgaatatgacgtattaagcgtcgggcattcatcaacctccatcagt gccggaattggtattgcggttgctgccgaaaaagaaggcaaaaatcgccgcaccgtctgtgtcattggcgatggcgcgattaccgcaggcatggcgtttg aagcgatgaatcacgcgggcgatatccgtcctgatatgctggtgattctcaacgacaatgaaatgtcgatttccgaaaatgtcggcgcgctcaacaacca tctggcacagctgctttccggtaagctttactcttcactgcgcgaaggcgggaaaaaagttttctctggcgtgccgccaattaaagagctgctcaaacgc accgaagaacatattaaaggcatggtagtgcctggcacgttgtttgaagagctgggctttaactacatcggcccggtggacggtcacgatgtgctggggc ttatcaccacgctaaagaacatgcgcgacctgaaaggcccgcagttcctgcatatcatgaccaaaaaaggtcgtggttatgaaccggcagaaaaagaccc gatcactttccacgccgtgcctaaatttgatccctccagcggttgtttgccgaaaagtagcggcggtttgccgagctattcaaaaatctttggcgactgg ttgtgcgaaacggcagcgaaagacaacaagctgatggcgattactccggcgatgcgtgaaggttccggcatggtcgagttttcacgtaaattcccggatc gctacttcgacgtggcaattgccgagcaacacgcggtgacctttgctgcgggtctggcgattggtgggtacaaacccattgtcgcgatttactccacttt cctgcaacgcgcctatgatcaggtgctgcatgacgtggcgattcaaaagcttccggtcctgttcgccatcgaccgcgcgggcattgttggtgctgacggt caaacccatcagggtgcttttgatctctcttacctgcgctgcataccggaaatggtcattatgaccccgagcgatgaaaacgaatgtcgccagatgctct ataccggctatcactataacgatggcccgtcagcggtgcgctacccgcgtggcaacgcggtcggcgtggaactgacgccgctggaaaaactaccaattgg caaaggcattgtgaagcgtcgtggcgagaaactggcgatccttaactttggtacgctgatgccagaagcggcgaaagtcgccgaatcgctgaacgccacg ctggtcgatatgcgttttgtgaaaccgcttgatgaagcgttaattctggaaatggccgccagccatgaagcgctggtcaccgtagaagaaaacgccatta tgggcggcgcaggcagcggcgtgaacgaagtgctgatggcccatcgtaaaccagtacccgtgctgaacattggcctgccggacttctttattccgcaagg aactcaggaagaaatgcgcgccgaactcggcctcgatgccgctggtatggaagccaaaatcaaggcctggctggcataataa
>BBa_K098995 Part-only sequence (935 bp)
tttatggctagctcagtcctaggtacaatgctagctactagagaaagaggagaaatactagatgagcacaaaaaagaaaccattaacacaagagcagctt
gaggacgcacgtcgccttaaagcaatttatgaaaaaaagaaaaatgaacttggcttatcccaggaatctgtcgcagacaagatggggatggggcagtcag
gcgttggtgctttatttaatggcatcaatgcattaaatgcttataacgccgcattgcttgcaaaaattctcaaagttagcgttgaagaatttagcccttc
aatcgccagagaaatctacgagatgtatgaagcggttagtatgcagccgtcacttagaagtgagtatgagtaccctgttttttctcatgttcaggcaggg
atgttctcacctgagcttagaacctttaccaaaggtgatgcggagagatgggtaagcacaaccaaaaaagccagtgattctgcattctggcttgaggttg
aaggtaattccatgaccgcaccaacaggctccaagccaagctttcctgacggaatgttaattctcgttgaccctgagcaggctgttgagccaggtgattt
ctgcatagccagacttgggggtgatgagtttaccttcaagaaactgatcagggatagcggtcaggtgtttttacaaccactaaacccacagtacccaatg
atcccatgcaatgagagttgttccgttgtggggaaagttatcgctagtcagtggcctgaagagacgtttggctgatactagagtcacactggctcacctt
cgggtgggcctttctgcgtttatatactagagagagaatataaaaagccagattattaatccggcttttttattattttactagagtaacaccgtgcgtg
ttgactattttacctctggcggtgataatggttgc
Summary for Week June 7th to June 11th
6-7-10:
a) Miniprep 2 pellet each for pBAD33, pBAD35, pBAD18, ygiV (attach to the cell wall), RcsB(expression factor that increase the clonic acid).
b) enzyme digestion for pBAD33, vgiV, pBAD18, and RcsB.
c) Gel extract for vgiV and RcsB and purification
d) PCR purification for pBAD18 and pBAD33
e) overnight ligation (set up by Sarah)
6-8-10:
a) transformation pBAD18-RcsB and pBAD33-vgiV into DH10B E. Coli. (Sarah and Hannah plates the cloning of pBAD18-RcsB and pBAD33-vgiV)
b) Miniprep For Encryption Project: BBa_I11020, BBa_K199021, BBa_K2902006, BBa_200021, BBa_B1006-A, BBa_B1006-B, BBa_E0240 For Drug Project: 2 pellets of each, pBAD18, pBAD33, pBAD35, vgiV, RcsB
6-9-10:
a) For the RcsB and VgiV plates, only a few colonies growed (they are really tiny, kindda hard to see, so we put it back to the incubator growing for couple of more hours)
b) Did the cloning of RcsB into pBAD18 and vgiV into pBAD33 again with different insert/vector ratio.
c) Plate the pellet and control.
6-10-10:
a) The pBAD18-RcsB plate has colonies growed.
b) Set up the screening with colony PCR
c) Made MRS Broth, MRS plates, no antibiotic plates, Amp+Kan plates, and CM plates with Mary.
 "
"