Team:WashU/Notebook/MolecularBiology
From 2010.igem.org

Week of 6/28
7/1
Plated The Megax BH10B strain and Strain 8 from the cohen lab onto an amp plate to check that the Megax BH10B strain is killed
Week of 7/12
7/13
Minipreped all the cultures and nanodropped them
Ran 3 PCR reactions (@ 58,60, and 62 C) with primers p1 and p2 in order to attach the kozak onto the YFP
Enzyme digested Nat (bglII and EcoRI), Kan (BamHI and EcoRI), and Promoter (EcoRI). Did 2 reactions each with ~500 ng DNA
7/14
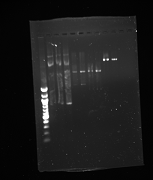
Gel from July 13th
Lanes from top to bottom:
1) Ladder 2) 5uL of 58 degree YFP + 1uL of loading dye 3) 5uL of 60 degree YFP + 1uL of loading dye 4) 5uL of 62 degree YFP + 1uL of loading dye 5) 5uL of KanMx4 1 + 1uL of loading dye 6) 5uL of KanMx4 2+ 1uL of loading dye 7) 5uL of NatMx4 1 + 1uL of loading dye 8) 5uL of NatMx4 2+ 1uL of loading dye 9) 5uL of Promoter 1 + 1uL of loading dye 10) 5uL of Promoter 2 + 1uL of loading dye
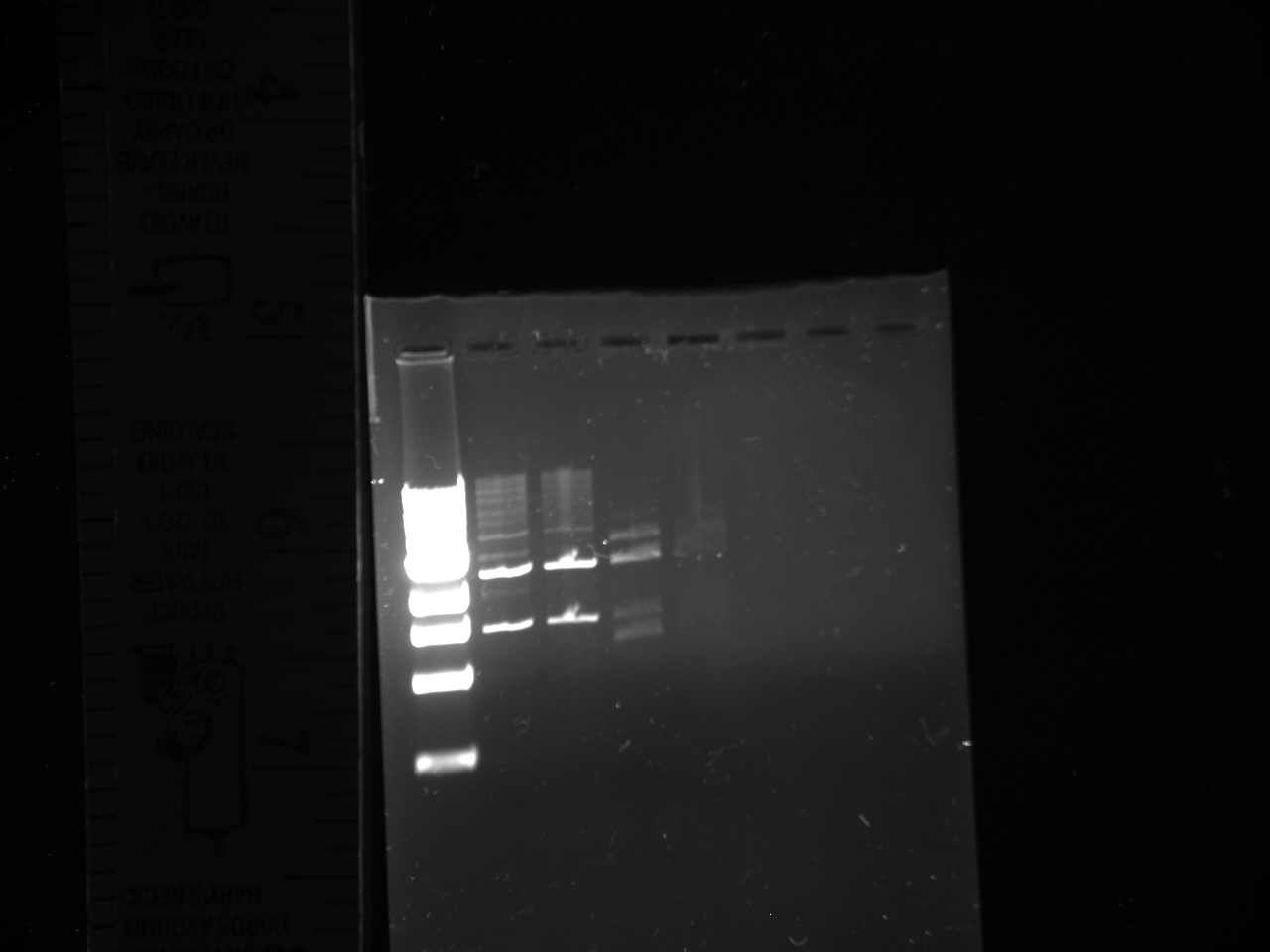
7/15
Gel reads from left to right as follows:
Dna Ladder; NatMX4-Promoter construct 2x; KanMX4 plasmid backbone 2x.
DNA ladder information is here at http://www.neb.com/nebecomm/products/productN3232.asp
7/16
Transformed EColi with the Kan plasmid containing the NatMX4 Promoter sequence. Bacteria Transformation July 16, 2010
1. Thaw competent cells on ice right before transformation. 2. Make 3 100uL tubes of competent cells 3. Add 4 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 250 ul room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 150 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
Week of 7/19
=7/19
Transformation of NatMX4/Promoter vector failed. Analysis of gel purification showed that the separate pieces which were ligated together on Thrusday July 15th were of the correct length. Conclusion: the ligation failed; most likely due to denatured ligase. The ligase icebath had melted prematurely and heat inactivated the enzyme. Yeast group obtained several replacement ligases and necessary buffers from the Cohen Laboratory in the 4444 building of the WashU Med School.
Transformation of C2 and SxL constructs into E. coli (AH)
-Reconstituted 5ug of each construct (in plasmid) in 10uL dH20. Final conc. of this stock solution is 500ng/uL. 10uL of working stock of each solution was made by diluting stock solution 1:10. Final conc. of working stock is 50ng/uL.
-Thaw 300uL tube of DH5alpha competent cells and divide among 5 tubes (60uL each). Amounts of 50ng/uL working stock were added to tubes labeled A-E in the following way:
A: 1uL C2 B: 5uL C2 C: 1uL SxL D: 5uL SxL E: Control=no DNA
-Incubate on ice 30 min, heat shock for 30s at 42C, rest on ice for 2 min. -Add 250uL of LB to each tube, Shake at 37C for 1hr -Plate 150uL on warm LB + Amp plates o/n at 37C
7/20
Digested 4 sequences:
1) NatMx4 with BglII and EcoRI: in microliters [5 buffer 3(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.]
2) Constitutive Promoter with EcoRI and SpeI: in uL [5 buffer 4(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.]
3) Construct 2 with BglII and XbaI: in uL [5 buffer 2(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of each enzyme; 41.5 of H2O.]
4) SxL with BamHI: in uL [5 buffer 3(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of BamHI; 41.5 of H2O.]
all were incubated for 4 hours followed 20 minutes of heat inactivation.
7/21
Digest SxL construct with EcoRI: added 1uL EcoRI to already done mix from 20 July digestion.
Gel purify NatMx4, Promoter, Construct 2, and KanMx4 digest results from 20 July 2010
Note: KanMx4 bands were not observed but approximate region of gel was removed for ligation KSB.
Ligate all three pieces together: NatMx4, Promoter, and Construct 2
2 reactions:
1) NCPA = NatMx4+Promoter+Construct 2
3.5uL Nat + 3.5uL Pro + 1.5uL C2
2) NCPB = NatMx4&Promoter+Construct 2 [NatMx4&Promoter was pre-existing ligation product from 15/16 Jul]
7uL NP + 1.5uL C2
Ligate KanMx4 with Sxl
2 reactions:
1) KSA = KanMx4 Digest 2 + SxL
7uL KanDigest2 + 1.5uL SxL
2) KSB = KanMx4 (purified) + SxL
7uL GelPureKanMx4 + 1.5uL SxL
Freezer stocks of C2 and SxL 1:1 ratio of LB was mixed with 100% glycerol to make a 50% glycerol/LB solution. 750uL of glycerol/LB solution was added to 750uL of each C2 and SxL culture from transformation on 7/19. Two tubes of each were placed at -80C.
7/22
Transformed the ligation DNA from reactions NCPA, NCPB, KSA, and KSB into E.Coli using the following protocol.
Bacteria Transformation
1. Thaw competent cells on ice right before transformation. 2. Add 4l of plasmid to cells, flick gently. 3. Incubate on ice 30 min. 4. Heat-shock cells for 30 seconds at 42oC w/o shaking 5. Transfer to ice, incubate 2 min 6. add 125 l room temp LB Media 7. Shake @ 37o for 1 hr + 8. Spread 100 l on a pre-warmed plate w/ correct antibiotic selector, allow sample to dry on plate 9. Incubate o/n at 37oC upside down
 "
"