|
|
| Line 1: |
Line 1: |
| - | {{Template:WITS-South_Africa_Main_Menu_19_07}}
| + | Final Machine 1 - LactoDetect |
| - | {{Template:WITS-South_Africa_Main_Menu_0910}}
| + | |
| | | | |
| | + | Construction: |
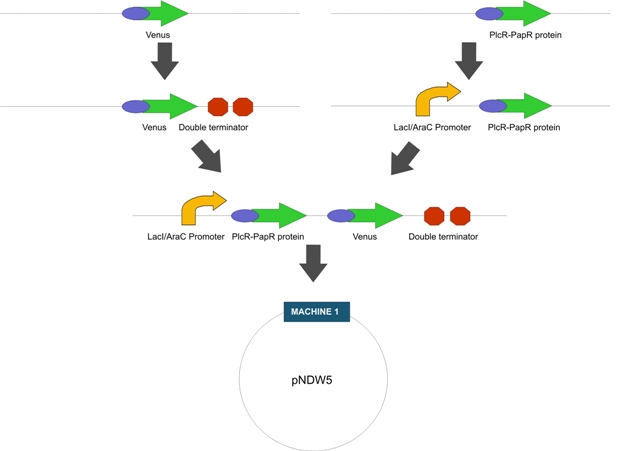
| | + | LactoDetect was constructed via a series of cloning steps using the Standard Assembly method. These are outlined in Figure 1 below. |
| | | | |
| - | | + | [[Image: machine_1_small.jpg|650px]] |
| - | == '''Cassette 2 in population 2''' ==
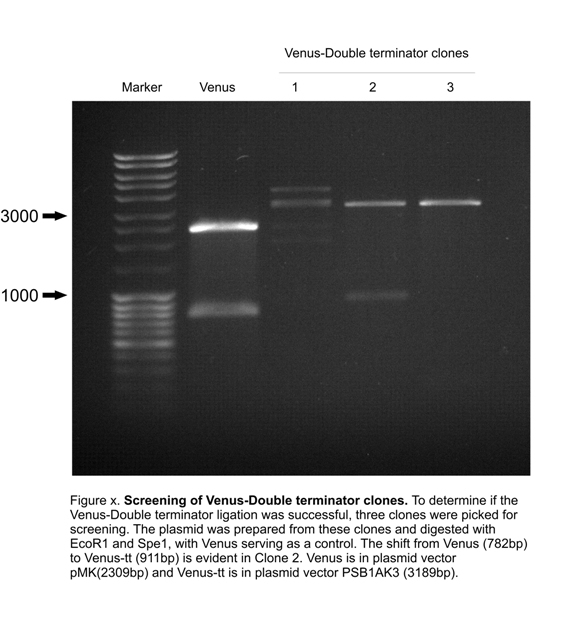
| + | [[Image:Venus-tt_screen.jpg|350px]] |
| - | | + | |
| - | </div>
| + | |
| - | | + | |
| - | [[Image:Cassette_2_wiki.JPG]] | + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | | + | |
| - | === PlcR Promoter ===
| + | |
| - | | + | |
| - | </div>
| + | |
| - | Through the identification of the PlcR box within the promoter of the PlcR regulon, a promoter that contains the PlcR box as well the requisite transcription-initiating landmarks within the promoter will be synthesized. Hence, any construct that is placed downstream of the promoter will be activated by the addition of exogenous PlcR and the facilitating peptide PapR. The PlcR-PapR promoter will be synthesized through primer-primer annealing and subsequent PCR elongation.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - |
| + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===PlcR-PapR Fusion Gene===
| + | |
| - | </div>
| + | |
| - | The ''plcR-papR'' fusion gene that codes for the activating peptides PlcR and PapR has been included in construct so as to amplify the initial signal presented by population 1. Since the fusion protein of PlcR and PapR is translated into its inactive form, there will be no direct self stimulatory effects within the same bacterium. Once secreted and processed, the peptides will serve to activate neighboring bacteria thus ensuring the propagation of the signal throughout the entire population.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===Phage Activator===
| + | |
| - | </div>
| + | |
| - | Developed by the winners of 2009 iGEM competition, Team Cambridge developed an array of activator and promoter combinations (derived from bacteriophages) that have varying degrees of binding and activating strengths. The intention was to create a library of activators and promoters than can be combined in such a way so as to vary the degree of negative feedback SpoOA exerts on the PlcR-PapR promoter. As such, several alternative endings of cassette 2 will be created, and based on experimental procedure a combination will be chosen so as to introduce an effective delay in repression. This delay is crucial so as to allow sufficient transcription of both the PlcR-PapR fusion peptide as well as the E. chromi pigment as this ensures adequate propagation of the signal throughout the colony as well as sufficient E. chromi being produced to ensure a distinctly visible report. The phage activator will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===mCherry===
| + | |
| - | </div>
| + | |
| - | Akin to the function of Venus in cassette 1, mCherry - a monomeric fluorescent protein - will be used in assaying the degree of transcription induced in colony 2 by incident PlcR-PapR quorum molecules. In addition to the visual report that will be produced as the response signal that propagates throughout the colony, mCherry will aid in the quantification of the mean fluorescent response of the colony. mCherry will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===Terminator===
| + | |
| - | </div>
| + | |
| - | As described above, the terminator ends transcription. By placing a terminator in the middle of cassette 2, it introduces a time delay between the activation of the initial region of cassette 2 and the feedback component, SpoOA. The terminator DNA will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===PO Phage Promoter===
| + | |
| - | </div>
| + | |
| - | Forming the receptor of the cognate activator-promoter pair of phage sensitivity tuners, the PO promoter will initiate transcription when the phage activator binds the promoter. Since the genes downstream of the promoter will only be transcribed after the phage activator is transcribed, translated and folded, a time delay is inserted due to this unavoidable metabolic process. The PO promoter will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===E. chromi===
| + | |
| - | </div>
| + | |
| - | So as to produce a chromogenic indication of the presence of HPV (or a proxy thereof), the E. chromi pigment will be used. The E. chromi biobrick is a composite biobrick comprised of sequential enzymes that process a substrate until a visible dye is produced. The combination of four sequential enzymes converts the substrate farnesyl pyrophosphate (which is colourless) to the pigment beta-carotine via several intermediates. The E. chromi biobrick will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===SpoOA===
| + | |
| - | </div>
| + | |
| - | So as to provide a suitable negative-feedback loop to the ‘machine’, SpoOA is a protein that binds to the regions adjacent to the PlcR box in the PlcR-PapR promoter. Once produced, the repression of SpoOA will negate the production of both mCherry as well as E. chromi. Furthermore, SpoOA will inhbit it’s own production this derepressing cassette 2 in such a way that the system, if left to its own devices in the presence of a suitable concentration of PlcR-PapR, will exhibit oscillatory behavior. SpoOA will be synthesized by Gene Art and will be codon optimized for ''Lactobacillus gasseri'' so as to ensure optimal production and expression.
| + | |
| - | | + | |
| - | | + | |
| - | ----
| + | |
| - | | + | |
| - | <div class="heading">
| + | |
| - | ===Terminator===
| + | |
| - | </div>
| + | |
| - | So as to provide a final cap to cassette 2 and the machine in whole, a terminator will be placed to end transcription. The terminator will be sourced from the Registry of Standard Parts.
| + | |
| - | | + | |
| - | | + | |
| - | </div>
| + | |
Construction:
LactoDetect was constructed via a series of cloning steps using the Standard Assembly method. These are outlined in Figure 1 below.
 "
"