Team:Uppsala-SwedenWeek9
From 2010.igem.org



Week-9
Identification of Bio-bricks
Plasmid of the samples which showed right size band on the gel were sent to a plasmid concentration test. We decided only F2, F6, F7, F8, F15 and F16 were in good concentration to proceed.
|
Sample |
A[260] |
A[280] |
Bg[320] |
Ratio |
Protein (μg/ml) |
Nucleic Acid (μg/ml) |
|
f2 |
0,0946 |
0,0607 |
0,0202 |
|
|
|
|
f3 |
0,0444 |
0,0346 |
0,0216 |
1,7474 |
2,9830 |
0,9640 |
|
f6 |
0,0844 |
0,0669 |
0,0300 |
1,4721 |
16,1731 |
2,0934 |
|
f7 |
0,1348 |
0,1020 |
0,0557 |
1,7074 |
11,9965 |
3,3075 |
|
f8 |
0,0905 |
0,0690 |
0,0420 |
1,7961 |
5,1758 |
2,0767 |
|
f11 |
0,0332 |
0,0261 |
0,0184 |
1,9261 |
0,7189 |
0,6559 |
|
f15 |
0,0769 |
0,0511 |
0,0205 |
1,8439 |
4,7573 |
2,4456 |
|
f16 |
0,0543 |
0,0389 |
0,0223 |
1,9290 |
1,5147 |
1,4182 |
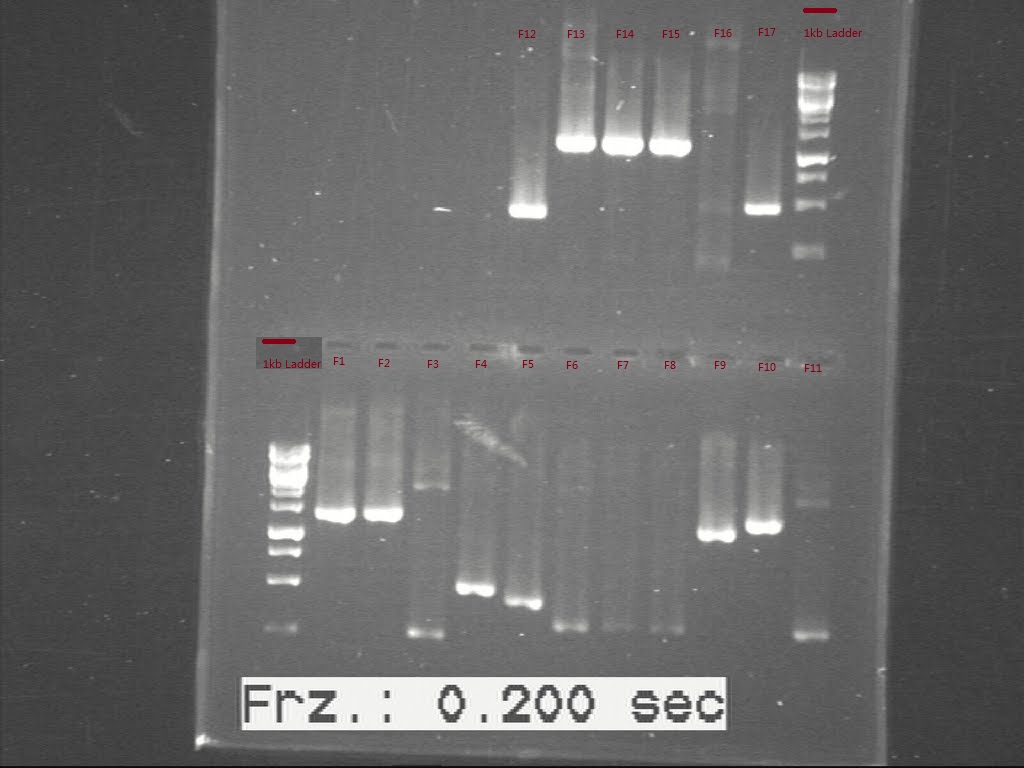
As the first PCR didn't show the length of all biobricks, the samples failed last time were sent to run another c-PCR. The result was showed in Fig.1
However, only sample F11 shows the band with right size on the gel pic. Samples, F1, F3, F4, F5, F9, F10, F11, F12, F13, F14 and F17 were inoculated again to get enough plasmid for further use. Second colony PCR was performed again for all the starting biobricks and the products were run for gel again. Comparing the DNA length to the band size, all the biobricks were confirmed, except for F16.
The concentration of plasmid were measured for all the samples and values were showed in Table 2
Analysis
Sample A[260] A[280] Bg[320] Ratio Protein Nucleic Acid
µg/ml µg/ml
____________________________________________________________________________________
f1 0,0183 0,0101 -0,0027 1,6428 3,9441 86,2531
f2 0,0477 0,0334 0,0160 1,8282 2,9083 137,1389
f3 0,0255 0,0132 -0,0021 1,8045 2,8374 118,5919
f4 0,0518 0,0398 0,0216 1,6573 5,4159 124,4569
f5 0,0108 0,0033 -0,0041 2,0017 0,2684 66,7742
f6 0,0137 0,0079 0,0006 1,7979 1,3839 56,0149
f7 0,0072 0,0010 -0,0071 1,7638 1,7529 60,7466
f8 -0,0035 -0,0080 -0,0151 1,6180 2,3362 47,0326
f9 0,0104 0,0035 -0,0054 1,7835 1,7833 67,4654
f10 0,0074 -0,0004 -0,0076 2,0846 -0,1927 68,6519
f11 0,0184 0,0088 -0,0063 1,6384 4,6986 101,2263
f12 0,0008 -0,0044 -0,0115 1,7349 1,6832 51,6802
f13 0,0157 0,0066 -0,0074 1,6499 4,2257 94,6587
f14 0,0033 -0,0031 -0,0100 1,9215 0,6691 58,6173
f15 0,0403 0,0345 0,0294 2,1530 -0,3966 50,2377
16 0,0182 0,0075 -0,0052 1,8335 2,0872 101,2592
f17 0,0082 0,0036 -0,0020 1,8332 0,9140 44,2838
Plasmid of F1, F2, F4, F9,F10,F12, F17,ccdb C3 and ccdb K3 were digested with proper enzymes bought from Sigma-Aldrich. The biobricks were ligated in pairs according to the plan by Quick Ligase kit from Fermentas. The ligation product were transformed into DH5α competent cells and each sample were plated on the agar plate which antibiotics matched to its backbone antibiotic resistance.
 "
"