Team:UTDallas/Results
From 2010.igem.org
(Difference between revisions)
(→Results) |
|||
| (3 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
===Results=== | ===Results=== | ||
| - | *All images were taken with Olympus IX81 automated inverted microscope specially equipped for live cell imaging. All | + | *All images were taken with Olympus IX81 automated inverted microscope specially equipped for live cell imaging. All experiments were performed with DH5α E.coli cells. The filter sets we used are: 545/30x (excitation) and 620/60m (emission) filters for DsRed, 470/40x (excitation) and 525/50m (emission) for GFP. Data collection and processing was performed by the SlideBook software. |
*The data was taken using cells that were grown overnight in 2mL of LB broth that had 50ug/mL of the appropriate antibiotic at 37C and 220rpm. This was used in a 1:50 dilution. Each sample had 40uL from the broth grown overnight and 2mL of the LB broth with the appropriate antibiotic. These were allowed to grow until a predetermined OD. Then the contaminants were added and the cells were allowed to grow for another 2 hours, before the images were taken. | *The data was taken using cells that were grown overnight in 2mL of LB broth that had 50ug/mL of the appropriate antibiotic at 37C and 220rpm. This was used in a 1:50 dilution. Each sample had 40uL from the broth grown overnight and 2mL of the LB broth with the appropriate antibiotic. These were allowed to grow until a predetermined OD. Then the contaminants were added and the cells were allowed to grow for another 2 hours, before the images were taken. | ||
| Line 10: | Line 10: | ||
[[Image:Ver2.jpg | 650 px | center]] | [[Image:Ver2.jpg | 650 px | center]] | ||
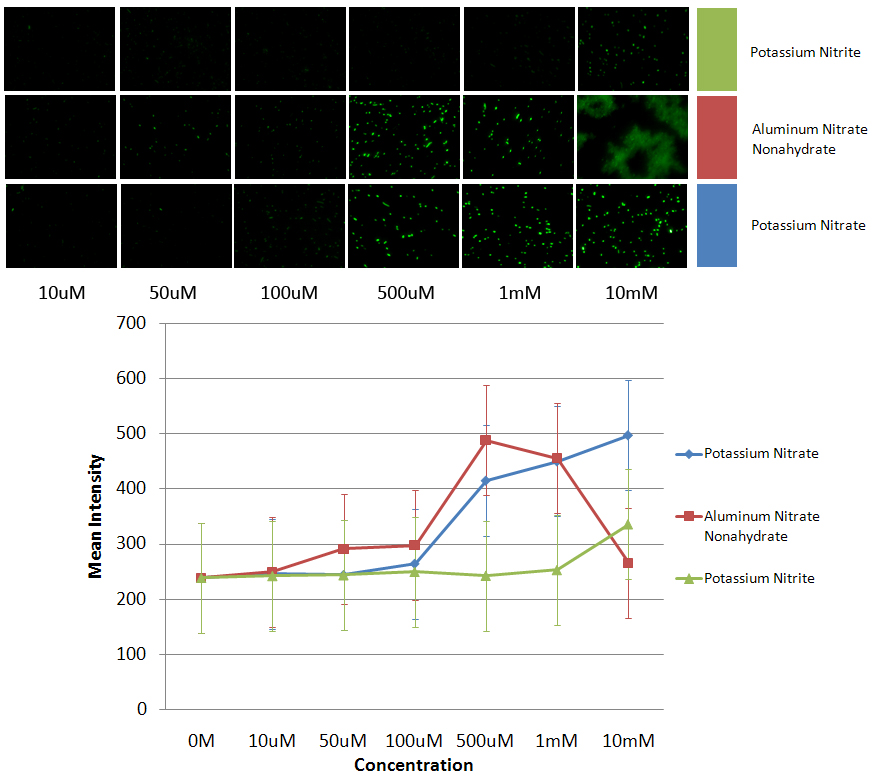
*The images shown are the images used to get the data. From left to right the image is the negative control to a 10mM concentration of the respective contaminants. The graph shows the average of all the maximum intensities of the cells in the image vs the concentration of nitrate/nitrite added to the sample. The data shows that PyeaR+GFP responds better to nitrates than nitrites. It also shows that the cell is more sensitive to aluminum nitrate nonahydrate, and therefore starts dying at a lower concentration. | *The images shown are the images used to get the data. From left to right the image is the negative control to a 10mM concentration of the respective contaminants. The graph shows the average of all the maximum intensities of the cells in the image vs the concentration of nitrate/nitrite added to the sample. The data shows that PyeaR+GFP responds better to nitrates than nitrites. It also shows that the cell is more sensitive to aluminum nitrate nonahydrate, and therefore starts dying at a lower concentration. | ||
| + | |||
| + | |||
| + | *PyeaR+GFP(BBa_K412000) Timelapse images | ||
| + | [[Image:tileview.jpg | 650 px | center]] | ||
| + | *The images above are timelapse images. The images were taken every ten minutes after the 10mM potassium nitrate was added to the sample. (It was not allowed to grow for two hours, like all the other images.) | ||
| + | |||
*PyeaR+RFP (BBa_K412001) | *PyeaR+RFP (BBa_K412001) | ||
[[Image:Pyear_RFP.jpg | 650 px | center]] | [[Image:Pyear_RFP.jpg | 650 px | center]] | ||
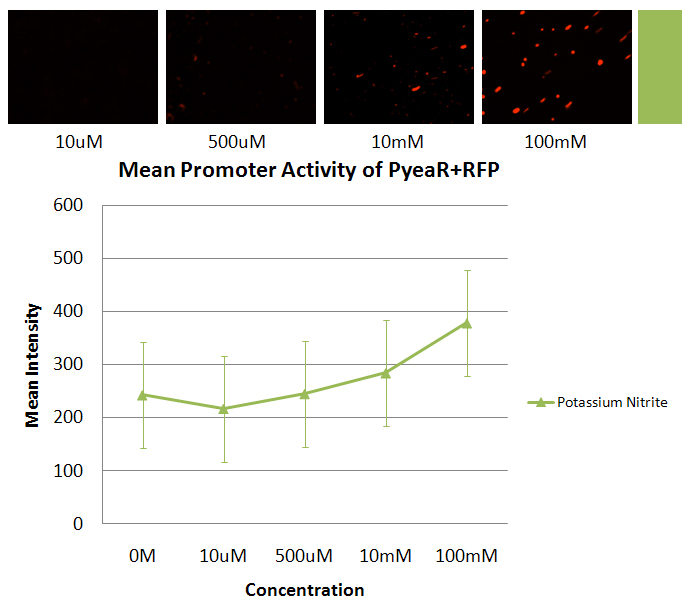
| - | *The images and the graph are from PyeaR+RFP incubated with potassium nitrite. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of nitrite added to the sample. | + | *The images and the graph are from PyeaR+RFP incubated with potassium nitrite. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of nitrite added to the sample. |
| + | |||
*Pu+GFP (BBa_K270003) | *Pu+GFP (BBa_K270003) | ||
| Line 19: | Line 26: | ||
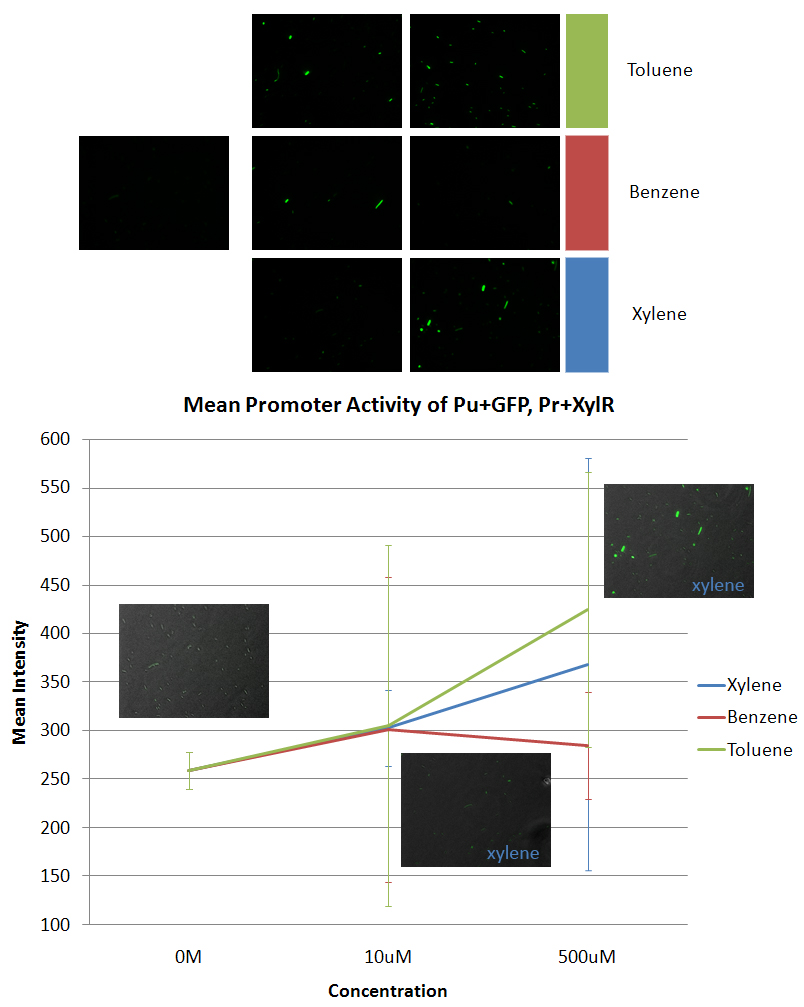
*The data was taken using E. coli DH5α that was co-transformed with Pu+GFP and Pr+XylR. | *The data was taken using E. coli DH5α that was co-transformed with Pu+GFP and Pr+XylR. | ||
*The leftmost image is the negative control. The other six images are the other concentrations (10uM and 500uM) in the specific aromatic: xylene, benzene, and toluene. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of the aromatic added to the sample. We also took data from 10mM and 100mM concentrations of aromatics, but most of the cells died at these higher concentrations. | *The leftmost image is the negative control. The other six images are the other concentrations (10uM and 500uM) in the specific aromatic: xylene, benzene, and toluene. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of the aromatic added to the sample. We also took data from 10mM and 100mM concentrations of aromatics, but most of the cells died at these higher concentrations. | ||
| + | |||
| + | |||
| + | *LacI+RFP, PyeaR+GFP (BBa_K412002) | ||
| + | [[Image:LacI+RFP, PyeaR+GFP.jpg | 650 px | center]] | ||
| + | *Preliminary measurements show that the PyeaR promoter does not work in combination with the LacI+RFP. The two right images is the sample incubated in 500uM potassium nitrite, and the two left images is the negative control. The LacI+RFP is constantly on (bottom images) while the PyeaR+GFP does not respond (top images). | ||
Latest revision as of 02:48, 28 October 2010
Results
- All images were taken with Olympus IX81 automated inverted microscope specially equipped for live cell imaging. All experiments were performed with DH5α E.coli cells. The filter sets we used are: 545/30x (excitation) and 620/60m (emission) filters for DsRed, 470/40x (excitation) and 525/50m (emission) for GFP. Data collection and processing was performed by the SlideBook software.
- The data was taken using cells that were grown overnight in 2mL of LB broth that had 50ug/mL of the appropriate antibiotic at 37C and 220rpm. This was used in a 1:50 dilution. Each sample had 40uL from the broth grown overnight and 2mL of the LB broth with the appropriate antibiotic. These were allowed to grow until a predetermined OD. Then the contaminants were added and the cells were allowed to grow for another 2 hours, before the images were taken.
- PyeaR+GFP (BBa_K412000)
- The images shown are the images used to get the data. From left to right the image is the negative control to a 10mM concentration of the respective contaminants. The graph shows the average of all the maximum intensities of the cells in the image vs the concentration of nitrate/nitrite added to the sample. The data shows that PyeaR+GFP responds better to nitrates than nitrites. It also shows that the cell is more sensitive to aluminum nitrate nonahydrate, and therefore starts dying at a lower concentration.
- PyeaR+GFP(BBa_K412000) Timelapse images
- The images above are timelapse images. The images were taken every ten minutes after the 10mM potassium nitrate was added to the sample. (It was not allowed to grow for two hours, like all the other images.)
- PyeaR+RFP (BBa_K412001)
- The images and the graph are from PyeaR+RFP incubated with potassium nitrite. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of nitrite added to the sample.
- Pu+GFP (BBa_K270003)
- The data was taken using E. coli DH5α that was co-transformed with Pu+GFP and Pr+XylR.
- The leftmost image is the negative control. The other six images are the other concentrations (10uM and 500uM) in the specific aromatic: xylene, benzene, and toluene. The graph is of the average of all the maximum intensities of the cells in the picture vs the concentration of the aromatic added to the sample. We also took data from 10mM and 100mM concentrations of aromatics, but most of the cells died at these higher concentrations.
- LacI+RFP, PyeaR+GFP (BBa_K412002)
- Preliminary measurements show that the PyeaR promoter does not work in combination with the LacI+RFP. The two right images is the sample incubated in 500uM potassium nitrite, and the two left images is the negative control. The LacI+RFP is constantly on (bottom images) while the PyeaR+GFP does not respond (top images).
 "
"