Team:UT-Tokyo/Sudoku assay LeakSw
From 2010.igem.org
(→Experiment 7) |
(→Detail protocols) |
||
| Line 181: | Line 181: | ||
== '''Detail protocols''' == | == '''Detail protocols''' == | ||
| + | |||
| + | '''Experiment7''' | ||
| + | |||
| + | materials<br/> | ||
| + | cre recombinase<br/> | ||
| + | x10 cre buffer<br/> | ||
| + | Hikari/pSB1C3…BBa_ concentration ng/ul / date:<br/> | ||
| + | onCMV plasmid 86ng/ul<br/> | ||
| + | 1%agarose gel<br/> | ||
| + | x10Mbuffer<br/> | ||
| + | XbaI<br/> | ||
| + | PstI<br/> | ||
| + | EcoRI<br/> | ||
| + | Promega Wizard SV Gel and PCR Clean-Up System<br/> | ||
| + | |||
| + | '''procedure'''<br/> | ||
| + | Linearization of plasmids<br/> | ||
| + | <br/> | ||
| + | mixing following reagents and incubation under 37℃<br/> | ||
| + | I. Hikari plasmid 3.8ul(ngDNA)<br/> | ||
| + | x10Mbuffer 3ul<br/> | ||
| + | XbaI 1ul<br/> | ||
| + | PstI 1ul<br/> | ||
| + | MilliQ 22.2ul<br/> | ||
| + | Total 30ul<br/> | ||
| + | II. onCMV plasmid 1ul(ngDNA)<br/> | ||
| + | x10Hbuffer 2ul<br/> | ||
| + | EcoRI 1ul<br/> | ||
| + | MilliQ 16ul<br/> | ||
| + | Total 20ul<br/> | ||
| + | <br/> | ||
| + | After incubated for several hours, 1uL of each was checked by 1% agarose gel electrophoresis. Since having got expected band patterns (following result session in detail), we purified DNA fragments under column purification, used 50 uL of Nuclease-free water for elution. We recorded UV absorbance spectrum and concentration. <br/> | ||
| + | <br/> | ||
| + | '''result''' | ||
| + | Following DNA fragments (purified) was extracted under above procedures. <br/> | ||
| + | Hikari XPcut fragments 9.4ng/ul<br/> | ||
| + | onCMV Ecut fragments 42.3ng/ul<br/> | ||
| + | |||
| + | Band patterns of electrophoresis<br/> | ||
| + | OnCMV-- 5300bp<br/> | ||
| + | Hikari-- 2000bp, 1150bp and faint 3100bp band (indicated one-cut fragments)<br/> | ||
| + | (no image)<br/> | ||
| + | |||
| + | These were used for next recombination process.<br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | '''Recombination procedure'''<br/> | ||
| + | <br/> | ||
| + | We mixed following solutions and incubated under 37℃<br/> | ||
| + | <br/> | ||
| + | Mix I Hikari-XPcut 3.8ul<br/> | ||
| + | x10cre buffer 3ul<br/> | ||
| + | cre recombinase 1ul<br/> | ||
| + | MilliQ 22.2ul<br/> | ||
| + | total 30ul<br/> | ||
| + | Mix II Hikari-XPcut 3.8ul<br/> | ||
| + | x10cre buffer 2ul<br/> | ||
| + | cre recombinase 2ul<br/> | ||
| + | MilliQ 21.2ul<br/> | ||
| + | Total 30ul<br/> | ||
| + | Mix III onCMV-Ecut 3.8ul<br/> | ||
| + | x10cre buffer 2ul<br/> | ||
| + | cre recombinase 2ul<br/> | ||
| + | MilliQ 22.2ul<br/> | ||
| + | total 30ul<br/> | ||
| + | After incubated for several hours, added x10loding dye and checked by electrophoresis<br/> | ||
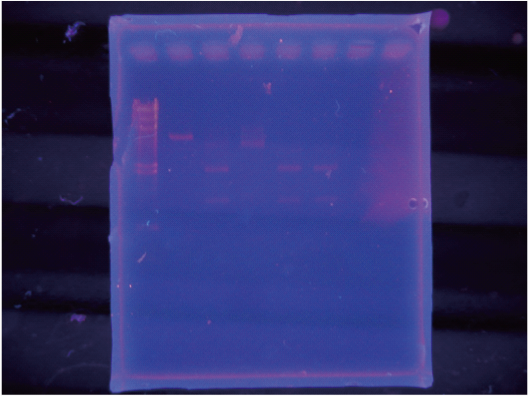
| + | The order of loading was:<br/> | ||
| + | lambda-Hmarker / onCMV-Ecut / 光XPcut / Mix I/ MixII/ MixIII<br /> | ||
{{UT-Tokyo_Foot}} | {{UT-Tokyo_Foot}} | ||
Revision as of 03:31, 28 October 2010


Sudoku
=>-Assay- Terminator Leak/ Location Sequence/ Phage MS2
Terminator Leak
Abstract
To construct 4C3 leak-switch, which is indispensable to determine the numbers when solving Sudoku, the following system is required: do not express enough amount of Cre gene that has two or more terminators on the upstream, and express enough amount of Cre gene that has only one terminator there. For this purpose, it is neccessary to use appropriate terminator.
The aim of terminator leakiness assay is to make sure whether our terminator is appropriate or not. We measured the fluorescence of GFP expressed when Cre protein recombine DNA construct. When the fluorescence of expressed GFP depends on the existence of Cre gene in this construct, our system works properly. However, in the first trial, the fluorescence of GFP cannot be observed in our construct. We considered whether lox sequence, cre coding sequence or both of them have some errors, and performed some experiments to make sure our hypothesis. As a result, it elucidated that lox sequence is not functional.
INDEX
(experiment1-4)
Profiling of the gene expression by T7 RNA polymerase
(experiment5,6)
Verification of the recombination activity of Cre protein
(experiment7)
Verification of the activity of the lox sequence in our construct
Experiment 1
Profiling of the gene expression by T7 RNA polymerase
First, we did some experiments to profile the gene expression by T7 RNA polymerase. The part 1-24 (T7 promoter – GFP unit) plasmid was transformed to BL21 (DE3) strain, and incubated on the LB plate at 37 oC.One colony was isolated and cultivated in two LB mediums. One sample was added IPTG (final conc. 1mM) and incubated at 37 oC. The samples were centrifuged and the supernatant were removed. We checked fluorescence from pellets on our eyes.
- DE3 strain is one of the E.coli strain which codes the gene of T7 RNA polymerase at the downstream of lac operon on its genome.
As a result, we could not observe the increasing of the GFP flourescence by addition of IPTG.The cultivated bacteria without the addition of IPTG turned green and those with the addition of IPTG did not turn green.
From this result, the following possibility was suggested. Green bacteria without the addition of IPTG → T7 RNA polymerase was expressed weakly and transcribed the mRNA coding gfp. White bacteria with the addition of IPTG → Overexpressed GFP made inclusion body.
Experiment 2
From the previous result, we expected that reduced IPTG concentration will prevent the formation of inclution body and performed experiment 1 in low IPTG concentration.We checked the GFP fluorescence of both samples on our eyes.
The GFP fluorescence of both samples were measured and there was no difference between the two samples .
It was suggested that BL21(DE3) is not proper to control the expression of gene expression.
We desided to use Rossetta (DE3) pLysS, that can repress the activity of the leaked T7 RNA polymerase.
Experiment 3
The plasmid containing part 1-24 was transformed to Rosetta (DE3) plysS.
Experiment 4
We improved the protocol to the following.
We used full growth samples preserved in glycerol made in experiment three. We added samples I-V into LB medium and cultured at 37oC. One colony was isolated and cultured in LB medium in duplicate. We added IPTG into one and cultured at room temperature. Then we observed green fluorescence. (The concentration of IPTG was 0.1mM)
As a result, we confirmed that IPTG is able to induce expression in samples I-III. Regarding IV, a small amount of green fluorescence was detected in the sample without IPTG too. We were not able to detect any green fluorescence in V whether IPTG was added or not.
Experiment 5
Verification of the recombination activity of Cre protein
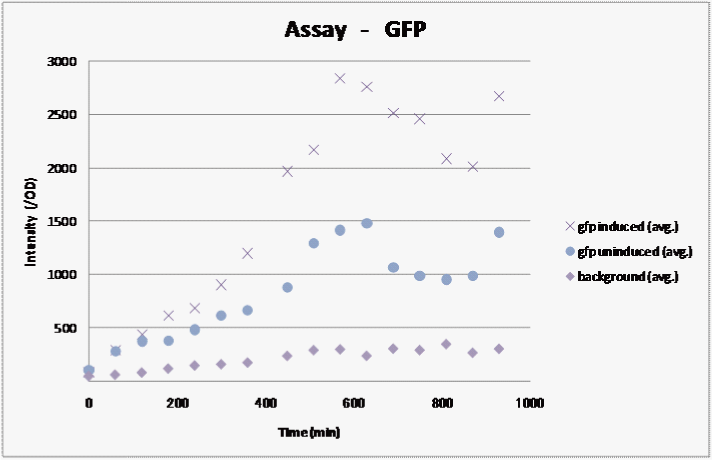
We assayed how the fluorescent intensity of GFP changes over time for sample 1, the sample we were able to induce in the previous assay..
I:The plasmid containing part 1-24 transformed to Rosetta (DE3) plysS
When the OD reached 0.6, we added IPTG to a final concentration of 0.1 mM.
We measured the fluorescence and graphed the results:
Result
We observed fluorescence without IPTG. However, fluorescence intensity was half compared to that after IPTG induction.
Experiment 6
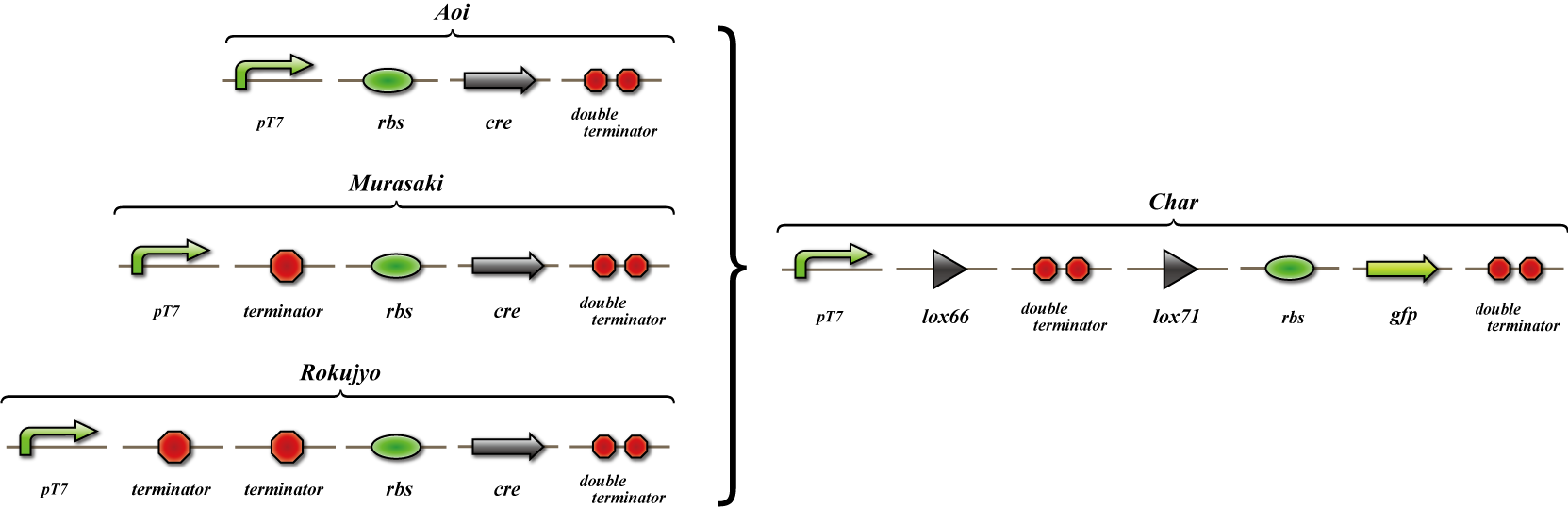
We make four constructs. The first, “Aoi,” does not have a terminator between the promoter and the ribosome binding site. The second, “Murasaki,” has one terminator, and “Rokujo” has two terminators. “Char,” the fourth, expresses GFP when the lox sites are recombined. We combine these to make three units. 1. Aoi + Char unit 2. Murasaki + Char unit 3. Rokujo + Char unit
Cre protein is placed downstream of the RBS.
The terminator is placed between the two lox sites. GFP is expressed when terminators between the lox sites are excised by the Cre protein.
GFP is expressed when the terminators between the lox sites are excised by Cre protein.
A terminator exists between the promoter and the rbs, so cre is not translated. However, this terminator is leaky and transcribes a small amount of cre, so the terminators are gradually excised and GFP is expressed.
Terminators exist between the promoter and the rbs, so cre is not translated. The terminator is not excised and GFP is not expressed.
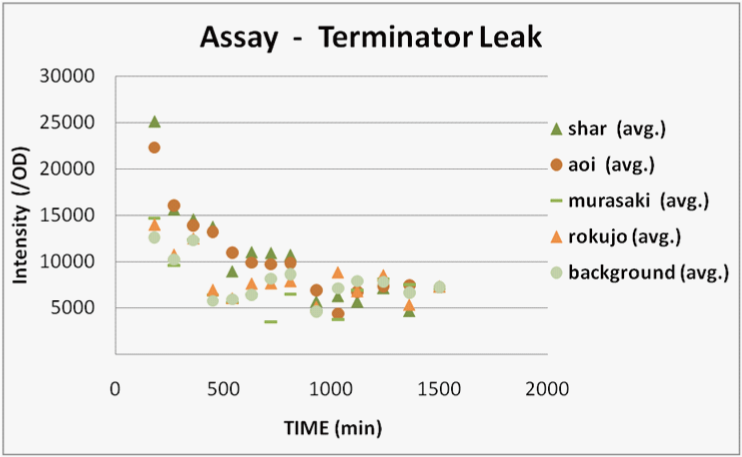
We transformed E.coli containing Char, Aoi&Char, Murasaki&Char and Rokujo&Char to Rosetta(DE3) plyS.We cultured these and measured the fluorescence intensity of GFP after IPTG induction.
With Char, we were not able to observe GFP fluorescence because terminator is not excised in this construct.(negative control)
With Murasaki & Char, IPTG induces the expression of Cre and the terminators are gradually excised, so the ratio of plasmids able to produce GFP increases.
Therefore, the intensity of GFP fluorescence increases gradually.Murasaki & Char express Cre at low levels, so the terminator is excised infrequently.Therefore, the intensity of GFP fluorescence increases slowly.
With Rokujo & Char, Cre is seldom expressed, so the intensity of GFP should be similar to that of Char.
Result
We were only able to observe fluorescence native to bacterial proteins. We deciphered this result as the following.
The sequence of our parts were identical to those reported on the registry, so the possibility of errors in our sequence was eliminated.
1. We used BBa_I71801(lox66)and (lox71)BBa_I718017 as our lox sites. There is the possibility that these sequences were dysfunctional.
2. We used BBa_J61047 as our Cre protein. There is the possibility that the sequence of this Cre protein was incorrect and Cre is dysfunctional.
Experiment 7
Verification of the activity of the lox sequence in our construct
We planned the following experiment to certify whether 1(in experiment 6) was true or not.
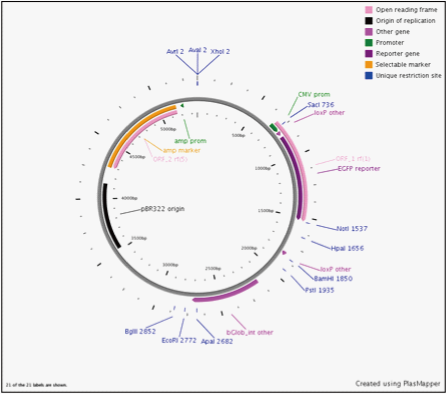
We dperformed experiments in vitro using purified the Cre protein. We enzymatically digested CMV and Char and adjusted the base length so that we could observe the base length alteration caused by Cre. CMV is a positive control previously reported to be recognized by Cre recombinase as a target of homologous recombination.
We cut this by Cre in vitro and observed the resulting band by electrophoresis.
As a result, we were able to confirm that Cre cuts on-CMV.
In contrast we were unable to observe a cut in the sample using Char.
Discussion
We concluded that lox66 and lox 71 incorporated into Char are dysfunctional or have very low excision efficiency.
Detail protocols
Experiment7
materials
cre recombinase
x10 cre buffer
Hikari/pSB1C3…BBa_ concentration ng/ul / date:
onCMV plasmid 86ng/ul
1%agarose gel
x10Mbuffer
XbaI
PstI
EcoRI
Promega Wizard SV Gel and PCR Clean-Up System
procedure
Linearization of plasmids
mixing following reagents and incubation under 37℃
I. Hikari plasmid 3.8ul(ngDNA)
x10Mbuffer 3ul
XbaI 1ul
PstI 1ul
MilliQ 22.2ul
Total 30ul
II. onCMV plasmid 1ul(ngDNA)
x10Hbuffer 2ul
EcoRI 1ul
MilliQ 16ul
Total 20ul
After incubated for several hours, 1uL of each was checked by 1% agarose gel electrophoresis. Since having got expected band patterns (following result session in detail), we purified DNA fragments under column purification, used 50 uL of Nuclease-free water for elution. We recorded UV absorbance spectrum and concentration.
result
Following DNA fragments (purified) was extracted under above procedures.
Hikari XPcut fragments 9.4ng/ul
onCMV Ecut fragments 42.3ng/ul
Band patterns of electrophoresis
OnCMV-- 5300bp
Hikari-- 2000bp, 1150bp and faint 3100bp band (indicated one-cut fragments)
(no image)
These were used for next recombination process.
Recombination procedure
We mixed following solutions and incubated under 37℃
Mix I Hikari-XPcut 3.8ul
x10cre buffer 3ul
cre recombinase 1ul
MilliQ 22.2ul
total 30ul
Mix II Hikari-XPcut 3.8ul
x10cre buffer 2ul
cre recombinase 2ul
MilliQ 21.2ul
Total 30ul
Mix III onCMV-Ecut 3.8ul
x10cre buffer 2ul
cre recombinase 2ul
MilliQ 22.2ul
total 30ul
After incubated for several hours, added x10loding dye and checked by electrophoresis
The order of loading was:
lambda-Hmarker / onCMV-Ecut / 光XPcut / Mix I/ MixII/ MixIII
 "
"