Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts
From 2010.igem.org
m (→BBa_K300003 - Phasin (PhaP) - internal domain) |
|||
| (19 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
<table width="100%" border="0"> | <table width="100%" border="0"> | ||
<tr> | <tr> | ||
| Line 28: | Line 29: | ||
</table> | </table> | ||
| + | |||
| + | =Improved Parts: list= | ||
| + | |||
| + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300002 - Phasin (PhaP) - head domain|BBa_K300002 - Phasin (PhaP) - head domain - improvement of BBa_K208001]] | ||
| + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300003 - Phasin (PhaP) - internal domain|BBa_K300003 - Phasin (PhaP) - internal domain - improvement of BBa_K208001]] | ||
| + | |||
| + | <br> | ||
| + | ---- | ||
| + | <br> | ||
</td></tr> | </td></tr> | ||
| Line 33: | Line 43: | ||
=Phasins= | =Phasins= | ||
| - | Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by | + | Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by several bacteria and that accumulate as intracellular granules. Phasins are proteins that can bind these granules. |
| - | + | The following parts are derived from the phaP gene of ''Ralstonia eutropha'', which encodes for a phasin, engineered without the stop codon in order to support protein fusions as a head/internal domain. | |
| - | + | ||
| - | In literature [Banki MR et al.] it has been shown that affinity tags composed by phasins assembled in tandem can increase the affinity with PHA. | + | Phasins can be used as affinity tags for a target protein, which can bind PHA granules allowing this way protein purification. |
| + | |||
| + | In literature [Banki MR et al., 2005] it has been shown that affinity tags composed by phasins assembled in tandem can increase the affinity with PHA. | ||
| + | |||
| + | The aim of our improvement of phasin already present in the Registry (<partinfo>BBa_K208001</partinfo>) was to provide a head and an internal protein domain useful to build fusion proteins. | ||
==<partinfo>BBa_K300002</partinfo> - Phasin (PhaP) - head domain== | ==<partinfo>BBa_K300002</partinfo> - Phasin (PhaP) - head domain== | ||
| - | This part can be used as a N-terminal affinity tag for a target protein that has to be fused downstream | + | This part can be used as a N-terminal affinity tag for a target protein that has to be fused downstream. Together with <partinfo>BBa_K300003</partinfo> enables the construction of composite tags. |
Because this part is a head domain, the Prefix is compatible with RFC10 and the Suffix is compatible with RFC23. | Because this part is a head domain, the Prefix is compatible with RFC10 and the Suffix is compatible with RFC23. | ||
| - | This part has been tested through <partinfo>BBa_K300086</partinfo> measurement system. | + | ===Construction=== |
| + | It is identical to <partinfo>BBa_K208001</partinfo>, but it lacks the stop codon in order to support protein fusions. | ||
| + | |||
| + | It has been designed as a head domain: | ||
| + | |||
| + | The Prefix sequence is 5'-GAATTCGCGGCCGCTTCTAG-3' (RFC10 Prefix) | ||
| + | |||
| + | The Suffix sequence is 5'-ACTAGTAGCGGCCGCTGCAG-3' (RFC23 Suffix) | ||
| + | |||
| + | For these reasons, a tail domain or an internal domain (compatible with RFC23) can be easily assembled downstream to create protein fusions. | ||
| + | |||
| + | To obtain this part, BioBrick <partinfo>BBa_K208001</partinfo> (provided by iGEM HQ in pSB3K3 in RFC23 standard) was PCR-amplified/mutagenized with primers: | ||
| + | |||
| + | phaP10F 5'-GCTTCTAGATGATCCTCACCCCGGAACA-3' | ||
| + | |||
| + | phaPSR: 5'-GCTACTAGTGGCAGCCGTCGTCTTCTTTG-3' | ||
| + | |||
| + | in order to delete the stop codon. | ||
| + | |||
| + | The PCR product was ran on a 1% agarose gel, gel-extracted, digested with XbaI-SpeI and ligated with <partinfo>pSB1AK3</partinfo> (previously cut with XbaI-SpeI and dephosphorylated). | ||
| + | Positive clones were selected through digestion screening/sequencing. | ||
| + | |||
| + | ---- | ||
| + | This part was used to construct the following synthetic composite affinity tags: | ||
| + | *<partinfo>BBa_K300093</partinfo> | ||
| + | *<partinfo>BBa_K300094</partinfo> | ||
| + | *<partinfo>BBa_K300095</partinfo> | ||
| + | *<partinfo>BBa_K300084</partinfo> | ||
| + | *<partinfo>BBa_K300097</partinfo> | ||
| + | |||
| + | or alone as a head domain; in this case it has been tested through <partinfo>BBa_K300086</partinfo> measurement system. | ||
| + | |||
===Methods=== | ===Methods=== | ||
Inoculum (into 5 ml LB+Amp) from glycerol stock of: | Inoculum (into 5 ml LB+Amp) from glycerol stock of: | ||
| - | *<partinfo>BBa_K300086</partinfo> | + | *<partinfo>BBa_K300086</partinfo>, |
| - | *<partinfo>BBa_K173000</partinfo> (positive control) | + | *<partinfo>BBa_K173000</partinfo> (positive control), |
| - | *<partinfo>BBa_B0031</partinfo> (negative control) | + | *<partinfo>BBa_B0031</partinfo> (negative control). |
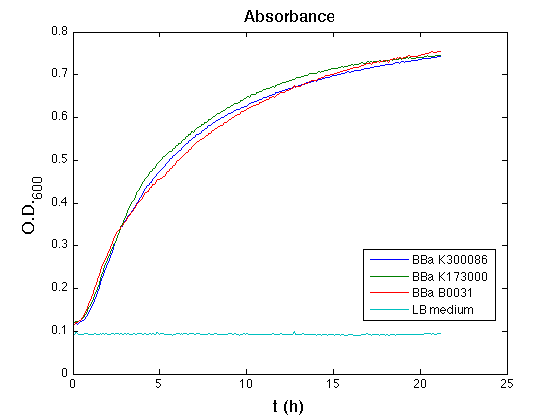
| - | + | ON cultures' growth at 37°C, 220 rpm. | |
| - | The following day cultures were diluted 1:100 and let | + | The following day, cultures were diluted 1:100 and let grown again for about five hours at 37°C, 220 rpm. |
| - | + | The optical density (O.D.) of each cell culture was than measured with TECAN Infinte F200. Samples were diluted in order to obtain the same O.D. equal to 0,02. | |
| - | + | Then we performed a 21-hours' experiment with measurements of absorbance and green fluorescence every five minutes using TECAN Infinite F200; cultures were shaken for 15 seconds every five minutes. Each value shown below is the mean of three measurements, from GFP data that of a non-fluorescent culture (negative control) was subtracted. | |
===Results=== | ===Results=== | ||
| Line 71: | Line 116: | ||
<table border="1" align="center"> | <table border="1" align="center"> | ||
<tr align="center"> | <tr align="center"> | ||
| - | <th>Culture</th><th>Doubling time [min.]</th> | + | <th>Culture</th><th>Doubling time [min.] ± std error</th> |
</tr> | </tr> | ||
<tr align="center"> | <tr align="center"> | ||
| - | <td><partinfo>BBa_K173000</partinfo></td><td> | + | <td><partinfo>BBa_K173000</partinfo></td><td>76.3336 ± 1.4362</td> |
</tr> | </tr> | ||
<tr align="center"> | <tr align="center"> | ||
| - | <td><partinfo>BBa_K300086</partinfo></td><td> | + | <td><partinfo>BBa_K300086</partinfo></td><td>73.6685 ± 1.6245</td> |
</tr> | </tr> | ||
<tr align="center"> | <tr align="center"> | ||
| - | <td><partinfo>BBa_B0031</partinfo></td><td> | + | <td><partinfo>BBa_B0031</partinfo></td><td>70.8421 ± 2.2181</td> |
</tr> | </tr> | ||
</table> | </table> | ||
===Discussion=== | ===Discussion=== | ||
| - | All cell cultures showed a similar growth curve | + | All cell cultures showed a similar growth curve; doubling time was computed as described [[Team:UNIPV-Pavia/Parts/Characterization#Doubling_time_evaluation|here]] in order to have informations about the burden due to the synthesis of such fusion proteins. It's possible to see that all doubling time are very similar; it's possible to assert that the expression of these BioBrick parts doesn't cause abnormal stress to cells. |
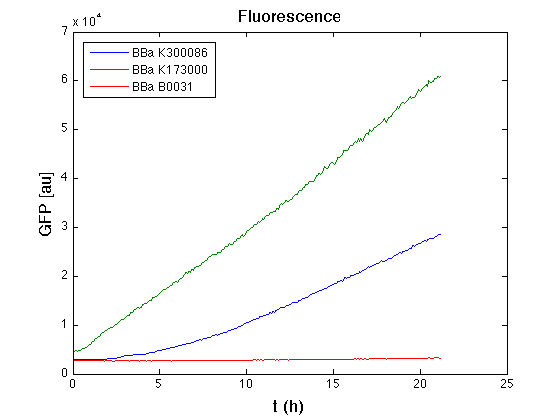
| - | + | From GFP curve it's possible to appreciate that in <partinfo>BBa_K300086</partinfo> GFP accumulation it's significantly different from that of negative control <partinfo>BBa_B0031</partinfo>. This result shows that the green fluorescent protein assembled downstream of the genetic circuit is correctly folded. | |
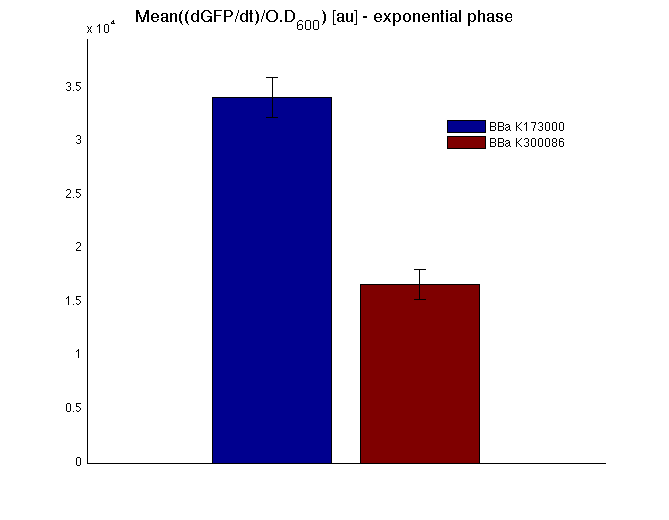
The mean protein synthesis rate was also computed over the growth exponential phase, showing an appreciable GFP production rate that is about a half of the positive control. | The mean protein synthesis rate was also computed over the growth exponential phase, showing an appreciable GFP production rate that is about a half of the positive control. | ||
==<partinfo>BBa_K300003</partinfo> - Phasin (PhaP) - internal domain== | ==<partinfo>BBa_K300003</partinfo> - Phasin (PhaP) - internal domain== | ||
| - | This part without stop codon and with Prefix and Suffix compatible with RFC23 (Silver Standard) in order to fully support protein fusions as an internal domain. Together with <partinfo>BBa_K300002</partinfo>, enables the construction of synthetic affinity tags | + | This part without stop codon and with Prefix and Suffix compatible with RFC23 (Silver Standard) was built in order to fully support protein fusions as an internal domain. Together with <partinfo>BBa_K300002</partinfo>, enables the construction of synthetic affinity tags containing phasins in tandem, possibly spaced by peptide linkers, as described in [Banki MR et al., 2005]. |
===Construction=== | ===Construction=== | ||
| + | It is identical to <partinfo>BBa_K208001</partinfo>, but it lacks the stop codon in order to support protein fusions. | ||
| + | |||
| + | It has been designed as an internal domain: | ||
| + | |||
| + | The Prefix sequence is 5'-GAATTCGCGGCCGCTTCTAG-3' (RFC23 Prefix) | ||
| + | |||
| + | The Suffix sequence is 5'-ACTAGTAGCGGCCGCTGCAG-3' (RFC23 Suffix) | ||
| + | |||
| + | For these reasons, a tail domain or an internal domain (compatible with RFC23) can be easily assembled downstream this BiobBrick to create protein fusions. | ||
| + | |||
| + | To obtain this part, BioBrick <partinfo>BBa_K208001</partinfo> (provided by iGEM HQ in pSB3K3 in RFC23 standard) was PCR-amplified/mutagenized with primers: | ||
| + | |||
| + | phaPSF: 5'-GCTTCTAGAATGATCCTCACCCCGGAACA-3' | ||
| + | |||
| + | phaPSR: 5'-GCTACTAGTGGCAGCCGTCGTCTTCTTTG-3' | ||
| + | |||
| + | in order to delete the stop codon. | ||
| + | |||
| + | The PCR product was ran on a 1% agarose gel, gel-extracted, digested with XbaI-SpeI and ligated with <partinfo>pSB1A3</partinfo> (previously cut with XbaI-SpeI and dephosphorylated). | ||
| + | Positive clones were selected through digestion screening/sequencing. | ||
| - | + | ---- | |
| + | This part was used as an internal domain to construct the following synthetic composite affinity tags: | ||
| + | *<partinfo>BBa_K300093</partinfo> | ||
| + | *<partinfo>BBa_K300094</partinfo> | ||
| + | *<partinfo>BBa_K300084</partinfo> | ||
| + | *<partinfo>BBa_K300097</partinfo> | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
Latest revision as of 03:25, 28 October 2010
|
|
|||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||
 "
"