Existing Parts from the Registry: list
<partinfo>BBa_K300009</partinfo>/<partinfo>BBa_I4102</partinfo> - PoPS->3OC6HSL sender device
<partinfo>BBa_F2620</partinfo> - 3OC6HSL -> PoPS Receiver
<partinfo>BBa_J61001</partinfo> - R6K Origin of replication
<partinfo>BBa_J23100</partinfo>, <partinfo>BBa_J23101</partinfo>, <partinfo>BBa_J23105</partinfo>, <partinfo>BBa_J23106</partinfo>, <partinfo>BBa_J23110</partinfo>, <partinfo>BBa_J23114</partinfo>, <partinfo>BBa_J23116</partinfo>, <partinfo>BBa_J23118</partinfo> - constitutive promoters from Anderson's collection
The <partinfo>BBa_J23100</partinfo>, <partinfo>BBa_J23101</partinfo>, <partinfo>BBa_J23105</partinfo>, <partinfo>BBa_J23106</partinfo>, <partinfo>BBa_J23110</partinfo>, <partinfo>BBa_J23114</partinfo>, <partinfo>BBa_J23116</partinfo>, <partinfo>BBa_J23118</partinfo> were charcterized in LB and M9 supplemented with glycerol (0.4%) growth media in high copy and low copy vectors in E. coli TOP10 (<partinfo>BBa_V1009</partinfo>).
RPU and doubling time were characterized for all of them, according to the protocols reported in this section.
The following measurement systems were used for high copy plasmids:
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23100</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23101</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23105</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23106</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23110</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23114</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23116</partinfo>
- <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23118</partinfo>
In order to build low copy plasmid measurement systems, the EcoRI-PstI fragment (J231xx-RFP) of each <partinfo>BBa_J61002</partinfo>-BBa_J231xx was assembled into <partinfo>pSB4C5</partinfo> vector. This fragment contains the constitutive promoter of interest upstream a RBS-RFP-TT expression system.
The following measurement parts were used for low copy plasmids:
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23100</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23101</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23105</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23106</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23110</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23114</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23116</partinfo>
- <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J23118</partinfo>
The RPU values and doubling times are here reported:
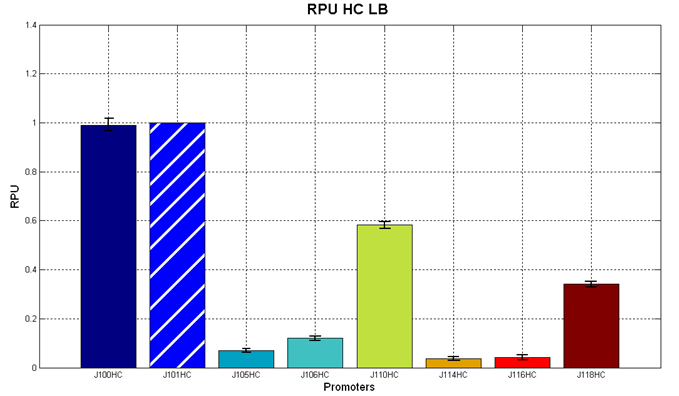 Figure 5 - R.P.U. of the studied promoters from Anderson promoters' collection, LB medium and high copy plasmid (<partinfo>BBa_J61002</partinfo>) | 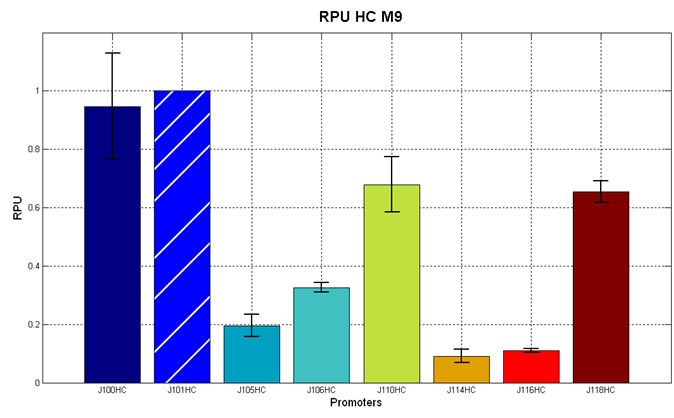 Figure 6 - R.P.U. of the studied promoters from Anderson promoters' collection, M9 medium and high copy plasmid (<partinfo>BBa_J61002</partinfo>) |
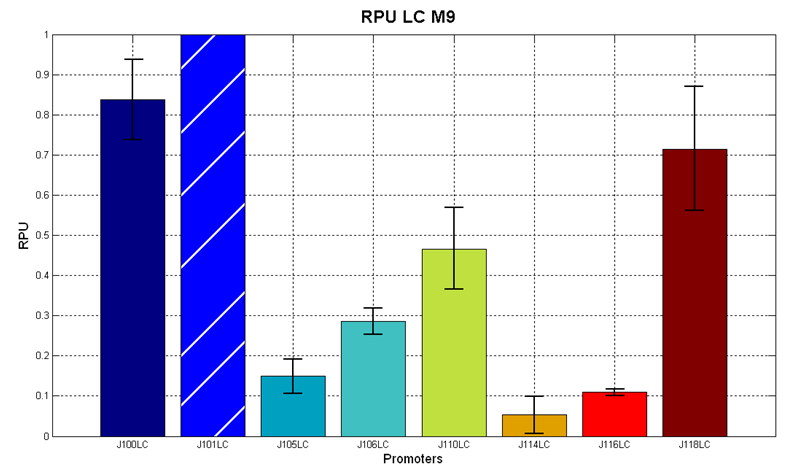 Figure 7 - R.P.U. of the studied promoters from Anderson promoters' collection, M9 medium and low copy plasmid (<partinfo>pSB4C5</partinfo>). These plasmids were constructed by assembling the EcoRI-PstI fragment of <partinfo>BBa_J61002</partinfo>-BBa_J231xx in <partinfo>pSB4C5</partinfo> vector, in order to transfer the promoter and the RBS-RFP-TT expression construct from <partinfo>BBa_J61002</partinfo> to <partinfo>pSB4C5</partinfo>. |
The error bars represent the standard deviation for three dfferent wells in the same experiment.
Doubling times were evaluated for the described cultures (HC stands for High Copy and LC stands for Low Copy):
| Promoter
| doubling time [minutes]
|
| LB in HC plasmid | M9 in HC plasmid | M9 in LC plasmid
|
| <partinfo>BBa_J23100</partinfo> | 33.75
±
1.34 | 82.53
±
2.45 | 86.11
±
4.45
|
| <partinfo>BBa_J23101</partinfo> | 35.93
±
0.62 | 82.68
±
1.84 | 86.42
±
1.91
|
| <partinfo>BBa_J23105</partinfo> | 29.86
±
0.33 | 63.09
±
7.08 | 85.00
±
5.13
|
| <partinfo>BBa_J23106</partinfo> | 29.17
±
0.96 | 68.11
±
4.25 | 88.71
±
0.90
|
| <partinfo>BBa_J23110</partinfo> | 31.28
±
0.42 | 67.52
±
5.87 | 76.15
±
2.16
|
| <partinfo>BBa_J23114</partinfo> | 28.97
±
0.49 | 59.44
±
5.20 | 80.12
±
0.95
|
| <partinfo>BBa_J23116</partinfo> | 28.14
±
0.25 | 72.74
±
0.37 | 81.68
±
3.08
|
| <partinfo>BBa_J23118</partinfo> | 32.84
±
0.31 | 73.64
±
2.41 | 89.86
±
2.93
|
It was not possible to evaluate promoters activities in low copy number plasmids in LB because the RFP activity was too weak and not distinguishable from the background.
Discussion: we observed that the ranking previously documented in the Registry is not valid in all the tested conditions, even if a general agreement can be observed. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking reported in the Registry.
<partinfo>BBa_P1004</partinfo> - chloramphenicol resistance cassette
<partinfo>BBa_K125500</partinfo> - GFP fusion brick
This part can be useful to construct fluorescent fusion proteins. It is composed by a tail domain (the GFP <partinfo>K125500</partinfo>) with a transcriptional terminator (<partinfo>BBa_B0015</partinfo>) downstream.
Other protein domains can be fused upstream of this part in order to create chimeric fluorescent proteins, or it can be ligated to tags useful for low-cost protein purification.
This part was used do design the following BioBrick measurement systems:
- <partinfo>BBa_K300086</partinfo>
- <partinfo>BBa_K300088</partinfo>
- <partinfo>BBa_K300090</partinfo>
- <partinfo>BBa_K300091</partinfo>
- <partinfo>BBa_K300092</partinfo>
- <partinfo>BBa_K300099</partinfo>
to test these contructs:
- <partinfo>BBa_K300002</partinfo>
- <partinfo>BBa_K300093</partinfo>
- <partinfo>BBa_K300094</partinfo>
- <partinfo>BBa_K300095</partinfo>
- <partinfo>BBa_K300084</partinfo>
- <partinfo>BBa_K300097</partinfo>
respectively. All of the tested parts are synthetic fusion tags whose activity could be measured by assembling a promoter with RBS upstream and a tail domain with terminator downstream, thus yielding the measurement systems. <partinfo>BBa_K300005</partinfo> was used as a tail domain with terminator downstream. It has been assembled to test the correct folding of the resulting fusion protein by measuring the GFP and to test the affinity tag performance with a proof of concept protein.
In all of the measurement parts, GFP could be successfully detected in bacteria harbouring the measurement parts in high copy plasmids. In this condition, GFP was detected by using an excitation filter at 485nm and an emission filter at 540nm in a Infinite F200 microplate reader (Tecan).
|
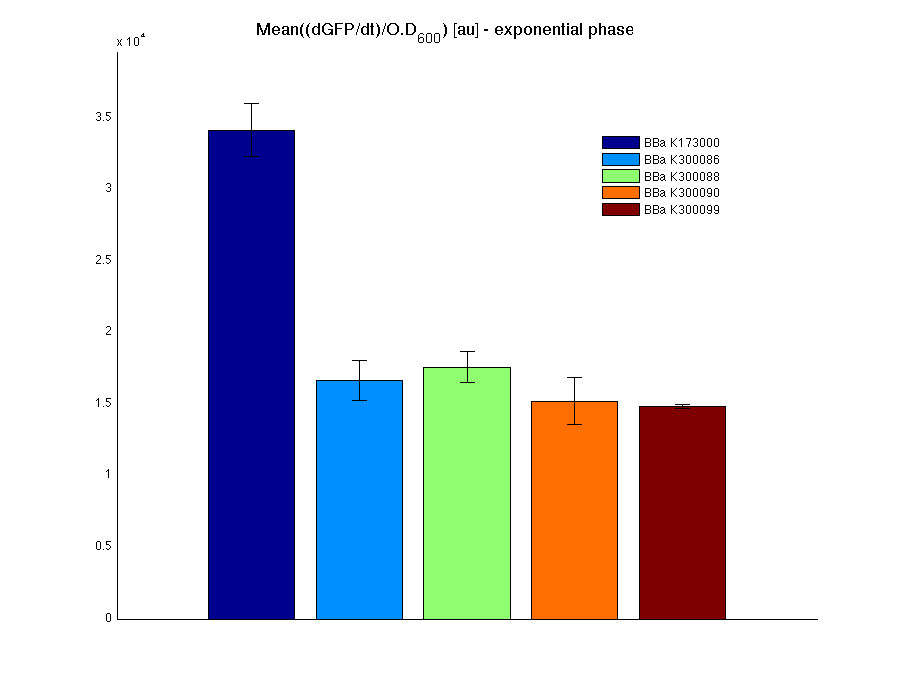 Mean (dGFP/dt)/O.D. over the exponential phase (under the hypothesis that GFP half-life in fusion contructs is similar to the original one - <partinfo>BBa_E0040</partinfo>) |
|
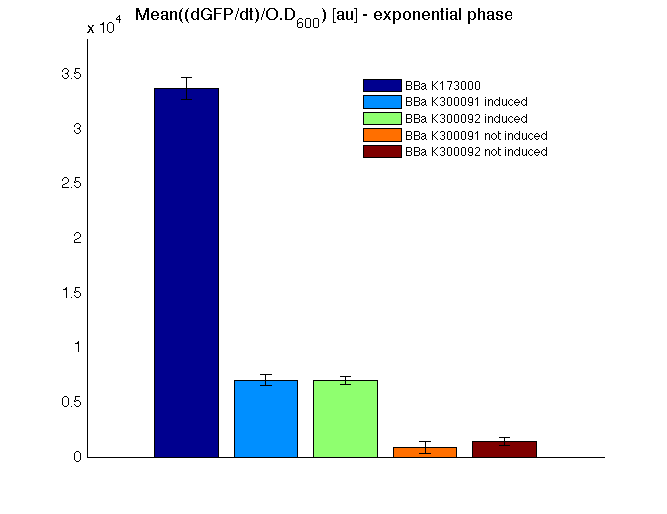 Mean (dGFP/dt)/O.D. over the exponential phase (under the hypothesis that GFP half-life in fusion contructs is similar to the original one - <partinfo>BBa_E0040</partinfo>) |
<partinfo>BBa_J72008</partinfo> - phi80 integration helper plasmid pInt80-649
|