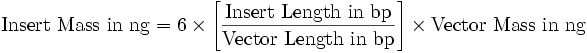Team:UNIPV-Pavia/Material Methods/Protocols
From 2010.igem.org
m (→PCR) |
|||
| (31 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
<!-- Contenuti --> | <!-- Contenuti --> | ||
| - | <td valign="top" | + | <td valign="top" > |
<table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | <table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | ||
<html><p align="center"><font size="4"><b>PROTOCOLS</b></font></p></html><hr><br><br> | <html><p align="center"><font size="4"><b>PROTOCOLS</b></font></p></html><hr><br><br> | ||
| Line 68: | Line 68: | ||
==M9 supplemented with glycerol (M9gly)== | ==M9 supplemented with glycerol (M9gly)== | ||
For 1L of medium, add: | For 1L of medium, add: | ||
| - | * 734 ml of autoclaved (and cooled to Tamb) ddH2O with a magnetic stirrer inside the bottle | + | * 734 ml of autoclaved (and cooled to Tamb, N.B. consider evaporation during autoclaving) ddH2O with a magnetic stirrer inside the bottle |
| + | * 5 ml of autoclaved 80% glycerol as carbon source | ||
* 100 ul of autoclaved or filtered (0.2um) CaCl2 1 M | * 100 ul of autoclaved or filtered (0.2um) CaCl2 1 M | ||
* 20 ml of 10% autoclaved casamino acids (dissolve 50 g in 500 ml = 10% stock) | * 20 ml of 10% autoclaved casamino acids (dissolve 50 g in 500 ml = 10% stock) | ||
| - | * 34 ml of filtered (0.2um) thiamine hydrochloride MW=337.27g/mol (340 mg in 34 ml) | + | * 34 ml of filtered (0.2um) thiamine hydrochloride MW=337.27g/mol (340 mg in 34 ml) (keep in mind it is photosensitive) |
* 2 ml of autoclaved MgSO4 1 M | * 2 ml of autoclaved MgSO4 1 M | ||
* 200 ml of autoclaved M9 salts 5x (dissolve 56.4 g in 1 liter ddH2O = 5x stock) | * 200 ml of autoclaved M9 salts 5x (dissolve 56.4 g in 1 liter ddH2O = 5x stock) | ||
| - | |||
* shake the ddH2O with the magnetic stirrer and start adding the other solutions in sterility (each solution must be completely dissolved!) in the order listed above. | * shake the ddH2O with the magnetic stirrer and start adding the other solutions in sterility (each solution must be completely dissolved!) in the order listed above. | ||
| - | * | + | * add antibiotic if needed |
* store at +4°C, protected from light | * store at +4°C, protected from light | ||
NOTE: | NOTE: | ||
* M9 salts 5x, 10% casamino acids, MgSO4 1 M and CaCl2 1 M can be stored at +4°C. | * M9 salts 5x, 10% casamino acids, MgSO4 1 M and CaCl2 1 M can be stored at +4°C. | ||
| - | * glycerol | + | * glycerol 80% can be stored at room temperature. |
| - | * thiamine hydrochloride (LIGHT SENSITIVE) is one-shot and must be prepared each time | + | * thiamine hydrochloride (LIGHT SENSITIVE) is one-shot and must be prepared each time (keep in mind you loose some volume during filtration) |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 164: | Line 164: | ||
::* adjust pH at 6.7 with KOH | ::* adjust pH at 6.7 with KOH | ||
::* 55mM (8.9 g/L) MnCl2 | ::* 55mM (8.9 g/L) MnCl2 | ||
| - | ::* filter (0.2 um) the solution and chill | + | ::* filter (0.2 um) the solution and chill in 50 ml |
: put the flask in ice when the culture reaches OD600=~0.05 (1mm pathlength – NanoDrop); | : put the flask in ice when the culture reaches OD600=~0.05 (1mm pathlength – NanoDrop); | ||
: aliquot in pre-chilled 50 ml falcon tubes; | : aliquot in pre-chilled 50 ml falcon tubes; | ||
| Line 172: | Line 172: | ||
: ICE: aliquot 100ul in pre-chilled 0.5ml tubes; | : ICE: aliquot 100ul in pre-chilled 0.5ml tubes; | ||
: put in -80°C freezer; | : put in -80°C freezer; | ||
| - | ALWAYS TEST THE EFFICIENCY IN [CFU/ug] UNITS | + | ALWAYS TEST THE EFFICIENCY IN [CFU/ug] UNITS: transform 100ul of competent cells with 4ng of DNA and 100ul of competent cells without DNA (add 1ul of ddH2O), then plate on proper LB agar plates. |
This protocol has shown to work with: | This protocol has shown to work with: | ||
| Line 181: | Line 181: | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
<br><br> | <br><br> | ||
| - | |||
==J. Sambrook, E.F. Fritsch, T. Maniatis (1989)== | ==J. Sambrook, E.F. Fritsch, T. Maniatis (1989)== | ||
| - | + | *DAY1 | |
| - | + | **Inoculum 5-8 ul from -80°C stock in 5 ml of LB (37°C, 220 rpm ON). | |
| - | + | *DAY2 | |
| - | + | **Dilution 1:500 in LB (flask, 30-37°C, 220 rpm), monitor OD600 until it reaches 0.04 (1mm pathlength – NanoDrop, it should take from 3 to 5 hours); | |
| - | + | **prepare the proper amount of 50 ml tubes in ice and pre-chill the centrifuge; | |
| - | + | **when the culture reaches the right OD600, aliquot the culture in the pre-chilled tubes; | |
| - | + | **centrifuge (4000rpm, 4°C, 10min) and discard the supernatant; | |
| - | + | **for each 50 ml of culture, add 30 ml of MgCl2-CaCl2 solution (Buffer1) and resuspend the pellet; | |
| - | + | **centrifuge (4000rpm, 4°C, 10min) and discard the supernatant; | |
| - | + | **for each 50 ml of the original culture, add 2 ml of CaCl2 solution (Buffer2) and resuspend the pellet; | |
| - | + | **aliquot in 0.5 ml tubes and store at -80°C. | |
ALWAYS TEST THE EFFICIENCY IN [CFU/ug] UNITS | ALWAYS TEST THE EFFICIENCY IN [CFU/ug] UNITS | ||
| + | |||
| + | ;Buffers preparation | ||
| + | *Buffer1: 80mM MgCl2, 20mM CaCl2 (e.g.: mix 8 ml MgCl2 1M, 2 ml CaCl2 1M and 90 ml ddH2O): | ||
| + | **put ddH2O into a flask or a bottle and autoclave it; | ||
| + | **add MgCl2 previously filter-sterilized (0,2 um) and CaCl2 previously autoclaved or filter-sterilized (0,2 um). | ||
| + | *Buffer2: 0.1 M CaCl2 and 15% of glycerol (e.g.: mix 100 mL of 1M CaCl2, 150 mL of 100% Glycerol and 750 mL of ddH2O): | ||
| + | **put ddH2O and glycerol into a flask and autoclave it; | ||
| + | **add CaCl2 previously autoclaved or filter-sterilized (0,2 um). | ||
| + | |||
| + | Keep Buffers cold. | ||
| + | |||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
<br><br> | <br><br> | ||
| Line 205: | Line 215: | ||
==TOP10== | ==TOP10== | ||
F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ- | F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ- | ||
| - | |||
* competent cells already prepared (5*10^7 CFU/ug with100ul of cells) | * competent cells already prepared (5*10^7 CFU/ug with100ul of cells) | ||
* competent cells from Invitrogen available (10^9 CFU/ug with 50ul of cells) | * competent cells from Invitrogen available (10^9 CFU/ug with 50ul of cells) | ||
| Line 218: | Line 227: | ||
==DH5alpha== | ==DH5alpha== | ||
F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK- mK+), λ– | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK- mK+), λ– | ||
| - | |||
* competent cells already prepared (10^8 CFU/ug with100ul of cells) | * competent cells already prepared (10^8 CFU/ug with100ul of cells) | ||
* commonly used for cloning | * commonly used for cloning | ||
| Line 225: | Line 233: | ||
==BW20767== | ==BW20767== | ||
F-, RP4-2(Km::Tn7,Tc::Mu-1), leu-163::IS10, ΔuidA3::pir+, recA1, endA1, thi-1, hsdR17, creC510 | F-, RP4-2(Km::Tn7,Tc::Mu-1), leu-163::IS10, ΔuidA3::pir+, recA1, endA1, thi-1, hsdR17, creC510 | ||
| - | |||
* competent cells already prepared (10^3 CFU/ug with100ul of cells) | * competent cells already prepared (10^3 CFU/ug with100ul of cells) | ||
* not used for cloning | * not used for cloning | ||
| Line 235: | Line 242: | ||
==XL1-Blue== | ==XL1-Blue== | ||
endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F'[ ::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK- mK+) | endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F'[ ::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK- mK+) | ||
| - | |||
* competent cells never prepared | * competent cells never prepared | ||
* a small stock of competent cells is available | * a small stock of competent cells is available | ||
| Line 244: | Line 250: | ||
==DB3.1== | ==DB3.1== | ||
F- gyrA462 endA1 glnV44 Δ(sr1-recA) mcrB mrr hsdS20(rB-, mB-) ara14 galK2 lacY1 proA2 rpsL20(Smr) xyl5 Δleu mtl1 | F- gyrA462 endA1 glnV44 Δ(sr1-recA) mcrB mrr hsdS20(rB-, mB-) ara14 galK2 lacY1 proA2 rpsL20(Smr) xyl5 Δleu mtl1 | ||
| - | |||
* competent cells already prepared (5*10^4 CFU/ug with 100ul of cells) | * competent cells already prepared (5*10^4 CFU/ug with 100ul of cells) | ||
* used for in vivo amplification of ccdB plasmids | * used for in vivo amplification of ccdB plasmids | ||
| Line 252: | Line 257: | ||
==STBL3== | ==STBL3== | ||
F- glnV44 recA13 mcrB mrr hsdS20(rB-, mB-) ara-14 galK2 lacY1 proA2 rpsL20 xyl-5 leu mtl-1 | F- glnV44 recA13 mcrB mrr hsdS20(rB-, mB-) ara-14 galK2 lacY1 proA2 rpsL20 xyl-5 leu mtl-1 | ||
| - | |||
* competent cells never prepared | * competent cells never prepared | ||
* used for in vivo amplification of DNA with direct repeats | * used for in vivo amplification of DNA with direct repeats | ||
| Line 260: | Line 264: | ||
==CW2553 + pJat8== | ==CW2553 + pJat8== | ||
Genotype: Khlebnikov A et al. (2000), Regulatable Arabinose-Inducible Gene Expression System with Consistent Control in All Cells of a Culture, Journal of Bacteriology, Vol. 182, No. 24, p.7029-7034. | Genotype: Khlebnikov A et al. (2000), Regulatable Arabinose-Inducible Gene Expression System with Consistent Control in All Cells of a Culture, Journal of Bacteriology, Vol. 182, No. 24, p.7029-7034. | ||
| - | |||
* pJat8 is Gentamycine resistant | * pJat8 is Gentamycine resistant | ||
NOTE: | NOTE: | ||
| Line 269: | Line 272: | ||
==MG1655 (seq)== | ==MG1655 (seq)== | ||
F-, λ-, rph-1 | F-, λ-, rph-1 | ||
| - | * | + | * CGSC#7740 |
* Fully sequenced genome (GenBank: NC_000913) | * Fully sequenced genome (GenBank: NC_000913) | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 275: | Line 278: | ||
==MC1061== | ==MC1061== | ||
F-, Δ(araA-leu)7697, [araD139]B/r, Δ(codB-lacI)3, galK16, galE15(GalS), λ-, e14-, mcrA0, relA1, rpsL150(strR), spoT1, mcrB1, hsdR2 | F-, Δ(araA-leu)7697, [araD139]B/r, Δ(codB-lacI)3, galK16, galE15(GalS), λ-, e14-, mcrA0, relA1, rpsL150(strR), spoT1, mcrB1, hsdR2 | ||
| - | * | + | * CGSC#6649 |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==BW25141== | ==BW25141== | ||
F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ-, galU95, ΔuidA3::pir+, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ-, galU95, ΔuidA3::pir+, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 | ||
| - | * | + | * CGSC#7635 |
* Carry the wild type pir gene | * Carry the wild type pir gene | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 286: | Line 289: | ||
==BW25142== | ==BW25142== | ||
F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ-, galU95, ΔuidA4::pir-116, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ-, galU95, ΔuidA4::pir-116, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 | ||
| - | * | + | * CGSC#6649 |
* Carry the pir-116 gene | * Carry the pir-116 gene | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 292: | Line 295: | ||
==BW23473== | ==BW23473== | ||
F-, Δ(argF-lac)169, ΔuidA3::pir+, recA1, rpoS396(Am)?, endA9(del-ins)::FRT?, rph-1, hsdR514, rob-1, creC510 | F-, Δ(argF-lac)169, ΔuidA3::pir+, recA1, rpoS396(Am)?, endA9(del-ins)::FRT?, rph-1, hsdR514, rob-1, creC510 | ||
| - | * | + | * CGSC#7837 |
* Carry the wild type pir gene | * Carry the wild type pir gene | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 298: | Line 301: | ||
==BW23474== | ==BW23474== | ||
F-, Δ(argF-lac)169, ΔuidA4::pir-116, recA1, rpoS396(Am)?, endA9(del-ins)::FRT, rph-1, hsdR514, rob-1, creC510 | F-, Δ(argF-lac)169, ΔuidA4::pir-116, recA1, rpoS396(Am)?, endA9(del-ins)::FRT, rph-1, hsdR514, rob-1, creC510 | ||
| - | * | + | * CGSC#7838 |
* Carry the pir-116 gene | * Carry the pir-116 gene | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 381: | Line 384: | ||
**0.4 µl dNTPs | **0.4 µl dNTPs | ||
**1 µl DNA (or ddH2O for blank sample). If you are performing a colony PCR, pick up the desired colony from a plate with a tip and dip it in the solution. | **1 µl DNA (or ddH2O for blank sample). If you are performing a colony PCR, pick up the desired colony from a plate with a tip and dip it in the solution. | ||
| - | **0. | + | **0.25 µl Taq Polymerase |
| - | ** | + | **0.5 ul VF2 primer (10 uM) |
| - | ** | + | **0.5 ul VR primer (10 uM) |
**A proper amount of ddH2O to have 20 µl of total reaction volume | **A proper amount of ddH2O to have 20 µl of total reaction volume | ||
*into an eppendorf tube. | *into an eppendorf tube. | ||
| Line 420: | Line 423: | ||
==1 kb Plus DNA Ladder preparation (Fermentas)== | ==1 kb Plus DNA Ladder preparation (Fermentas)== | ||
Mix gently: | Mix gently: | ||
| - | *1ul of DNA ladder | + | *1ul of DNA ladder (1 kb) |
*1ul of 6X DNA Loading Dye | *1ul of 6X DNA Loading Dye | ||
*4ul of Deionizied water | *4ul of Deionizied water | ||
| Line 437: | Line 440: | ||
=X-Gal staining protocol for beta galactosidase (blue/white screening)= | =X-Gal staining protocol for beta galactosidase (blue/white screening)= | ||
*The principle is that X-Gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) turns blue when reacts with beta-galactosidase. | *The principle is that X-Gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) turns blue when reacts with beta-galactosidase. | ||
| - | *Mix 20 ul X-Gal | + | *Mix 20 ul X-Gal 40 mg/ml and 60 ul LB and spread on required LB agar plates (X-Gal and DMF are toxic, use face-mask for your safety!!! X-Gal is light-sensitive, remember to keep it in the dark, when possible). |
| - | * | + | *If you have to induce beta-galactosidase production (for example in lac operon) add 20 ul of IPTG 200 mM to the mix. |
| - | *Result: blue colonies express LacZ, while white colonies don't | + | *Let plates dry at 37°C and than plate bacteria. |
| + | *Incubate at 37°C. | ||
| + | *Result: blue colonies express LacZ, while white colonies don't. | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | <br/><br/> | ||
=Sudan Black staining protocol= | =Sudan Black staining protocol= | ||
| - | |||
| - | |||
*Take 70 ul of colture and spread it onto a slide. | *Take 70 ul of colture and spread it onto a slide. | ||
*Allow the smear on the slide to dry. | *Allow the smear on the slide to dry. | ||
| Line 454: | Line 459: | ||
*When the slide is completely dry, add a drop of immersion oil directly to the slide. | *When the slide is completely dry, add a drop of immersion oil directly to the slide. | ||
*Examine the slide with optical microscope with 100x oil immersion objective. | *Examine the slide with optical microscope with 100x oil immersion objective. | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | <br/><br/> | ||
| + | |||
| + | =Yeast cultures= | ||
| + | |||
| + | ==Liquid YPD medium (0.5 L)== | ||
| + | |||
| + | *5 g yeast extract | ||
| + | *10 g peptone | ||
| + | *450 ml ddH2O | ||
| + | *Autoclave | ||
| + | *Add 50 ml of 20% glucose to reach the final concentration of 2% | ||
| + | *Add 2 ml of G418 geneticin (50 mg/ml stock) at the final concentration of 200 ug/ml | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==YPD agar (0.5 L)== | ||
| + | |||
| + | *5 g yeast extract | ||
| + | *10 g peptone | ||
| + | *10 g agar | ||
| + | *450 ml ddH2O | ||
| + | *Autoclave | ||
| + | *Add 50 ml of 20% glucose to reach the final concentration of 2% | ||
| + | *Add 2 ml of G418 geneticin (50 mg/ml stock) at the final concentration of 200 ug/ml | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==G418 stock solution (50 mg/ml)== | ||
| + | |||
| + | *Add 20 ml of ddH2O to 1 g of G418 powder (Sigma) | ||
| + | *Filter-sterilize (0.2 um) and aliquot in 1 ml stocks | ||
| + | *Store at +4°C as recommended by Sigma | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==LiAc 1M== | ||
| + | |||
| + | *Dissolve 1 g of LiAc dihydrate (Sigma) in ddH2O to a final volume of 10 ml | ||
| + | *Filter-sterilize (0.2 um) and store at +4°C | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==PEG 3350 50%== | ||
| + | |||
| + | *Dissolve 20 g of PEG 3350 (Sigma) in ddH2O on a magnetic stirrer to a final volume of 40 ml | ||
| + | *Autoclave | ||
| + | *Store at room temperature | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==Long term glycerol stocks== | ||
| + | |||
| + | *Mix 810 ul of an overnight yeast culture with 190 ul of sterile 80% glycerol | ||
| + | *Vortex and store at -80°C | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | ==Yeast transformation== | ||
| + | *Follow this protocol: http://openwetware.org/wiki/High_Efficiency_Transformation until “Pipette | ||
| + | 1.0 ml of sterile water into each tube; stir the pellet by with a micropipette tip and vortex” step. | ||
| + | *Centrifuge at 3000g 5 min, remove the supernatant and inoculate the pellet in 1 ml of pre-warmed | ||
| + | YPD in a 15 ml falcon tube. | ||
| + | *Incubate the cultures at 30°C, 200rpm for 2h. | ||
| + | *Centrifuge at 3000g 5 min, remove the supernatant and resuspend the pellet in the remaining | ||
| + | YPD. | ||
| + | *Plate the whole cells on a G418 plate, pre-incubated at room temperature. | ||
| + | *Incubate the plated cells at 28-30°C until colonies appear. | ||
Latest revision as of 07:11, 26 May 2011
|
|
||||||||||||||
|
|
|||||||||||||
 "
"