Team:UIUC-Illinois-Software/Project
From 2010.igem.org
Sordfysh48 (Talk | contribs) (added lots of description) |
Sordfysh48 (Talk | contribs) |
||
| Line 14: | Line 14: | ||
==BioMORTAR 3-Part Design Suite== | ==BioMORTAR 3-Part Design Suite== | ||
<br/> | <br/> | ||
| + | [[Image:BioMORTAR_Flowchart.png|right|500px]] | ||
===IMPtools=== | ===IMPtools=== | ||
*User inputs the desired starting and ending compounds | *User inputs the desired starting and ending compounds | ||
Revision as of 20:35, 27 October 2010
 | ||
Contents |
BioMORTAR Project Abstract
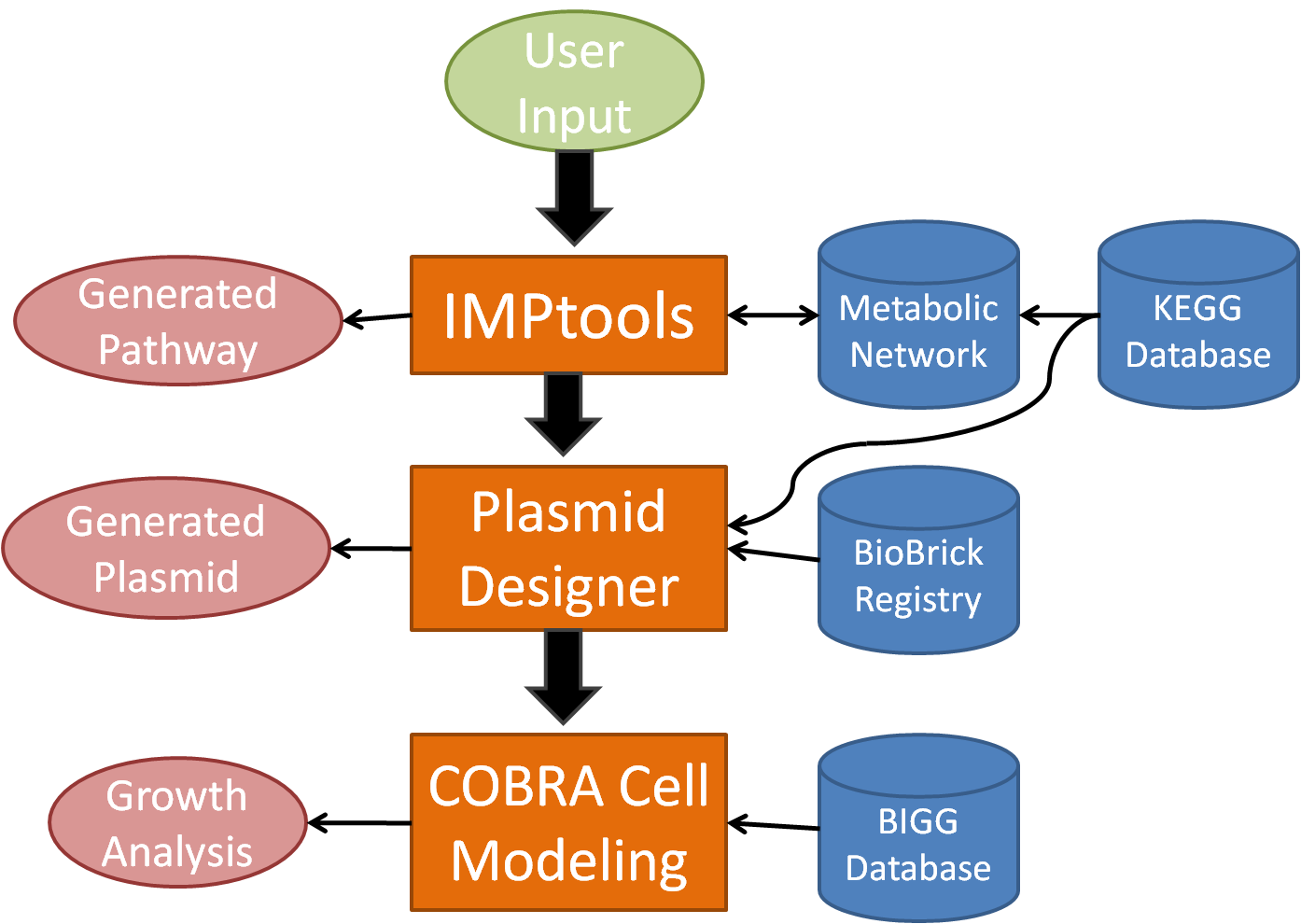
In order to facilitate the design process for novel bacterial metabolism, our team has created a tool suite known as BioMortar which will automate plasmid design for metabolic processes as well as model cell growth. BioMortar begins with a much improved version of IMPtools, which uses an algorithm over a network generated by the KEGG database, to determine the optimal metabolic pathway according to specified conditions. At this point, it accesses the DNA sequences for each recommended enzyme for each reaction and searches the BioBrick database for related gene sequences. Then, BioMORTAR designs and displays the advised, usable plasmid(s) in BioBrick format for the user. Finally, the program models the growth of the organism, with the addition of the new metabolic pathway(s). By automating the design process, BioMORTAR streamlines the process of designing bacteria with new metabolic processes.
Project Description
The 2010 Illinois iGEM Tools Team is in the phase of constructing a program to design a plasmid in-silico for the user-specified host bacteria as well as model the growth of the designed cell.
The web-based program will begin by implementing the desired metabolic pathway computed by a greatly improved version of last year's project, IMPtools, then by determining the recommended enzymes for each reaction, and accessing the DNA sequence for it. In the meantime, the program will model the growth of the user specified bacteria with the new metabolic pathway to optimize the plasmid design. Finally, it will construct and display, with annotation, the advised plasmid(s) for the user as well as display important information on projected growth. In addition, it will have the option to adhere to the iGEM biobrick standards and will reference the biobrick database.
As a whole, the program will not only give the user the optimal plasmid design for the metabolic process, but it will project growth for the modified bacteria and connect the user with the BioBrick database which can reference the necessary plasmids.
BioMORTAR 3-Part Design Suite
IMPtools
- User inputs the desired starting and ending compounds
- Weighting system allows for a more customized metabolic pathway
- IMPtools selects the best metabolic pathway from the generated reaction network
- An option allows IMPtools to optimize output while minimizing intermediate inputs or products through pathway branching
Plasmid Designer
- Reads the enzymes from the generated metabolic pathway
- Searches the databases for the genes for the required enzymes
- Attaches specified promoters, ribosome binding sites, and terminators
- Organizes all of the genetic code and creates a plasmid in silico
- References relevant genes on various databases, allowing the user to easily create the plasmid in the lab
COBRA cell Modeling
- Calculates the growth rate and robustness of bacteria with the added metabolic pathway
- Determines the feasibility of survival with the new reactions
- May be able to optimize the plasmid for survivability
Judging Criteria
The requirements to earn a Bronze Mousepad are:
- Register the team, have a great summer, and have fun attending the Jamboree.
- Create and share a Description of the team's project via the iGEM wiki.
- Present a Poster and Talk at the iGEM Jamboree.
- Develop and make available via the Registry an open source software tool that supports synthetic biology based on BioBrick standard biological parts (remember, the iGEM judges will be looking for substantial team-based software projects).
The requirements to earn a Silver Mousepad, in addition to the Bronze Mousepad requirements, are:
- Provide a detailed, draft specification for the next version of your software tool, or a second, distinct software tools project.
The requirements to earn a Gold Mousepad, in addition to the Silver Mousepad requirements, are:
- Help another iGEM team by, for example, analyzing a Part, debugging a Device, or modeling or simulating a System.
- Develop and document a new technical standard that supports the (i) design of BioBrick Parts or Devices, or (ii) construction of BioBrick Parts or Devices, or (iii) characterization of BioBrick Parts or Devices, or (iv) analysis, modeling, and simulation of BioBrick Parts or Devices, or (v) sharing of BioBrick Parts or Devices, either via physical DNA or as information via the internet.
- Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
 "
"